
中国水稻科学 ›› 2016, Vol. 30 ›› Issue (2): 111-120.DOI: 10.16819/j.1001-7216.2016.5138
• • 下一篇
孟姣1, 王海华1,2,3,*( ), 向建华1, 蒋丹1, 彭喜旭1,3, 贺欢欢1
), 向建华1, 蒋丹1, 彭喜旭1,3, 贺欢欢1
收稿日期:2015-09-16
修回日期:2015-11-30
出版日期:2016-03-10
发布日期:2016-03-10
通讯作者:
王海华
基金资助:
Jiao MENG1, Hai-hua WANG1,2,3,*( ), Jian-hua XIANG1, Dan JIANG1, Xi-xu PENG1,3, Huan-huan HE1
), Jian-hua XIANG1, Dan JIANG1, Xi-xu PENG1,3, Huan-huan HE1
Received:2015-09-16
Revised:2015-11-30
Online:2016-03-10
Published:2016-03-10
Contact:
Hai-hua WANG
摘要:
一氧化氮(nitric oxide, NO)信号分子与WRKY转录因子均参与植物抗逆、发育与代谢等许多生理过程。采用Agilent水稻全基因组cDNA芯片分析了NO处理后1、6和12 h水稻幼苗WRKY转录因子基因的表达谱,鉴定出在1个时间点有两倍或两倍以上表达变化的WRKY基因32个,主要分布在WRKY的Ⅰ和Ⅱ组,其中75%的Ⅱa和45.6%的Ⅱd亚组成员为差异表达基因;鉴定出至少在2个时间点有两倍或两倍以上表达变化的WRKY基因15个,均为早期(1 h)应答,且多数(64.2%)持续上调;基因功能预测分析表明,这些基因主要参与生物学过程中的细胞过程、代谢过程和刺激响应,以及分子功能中的转录调节活性和结合;代谢通路分析表明,WRKY24涉及植物与病原菌相互作用代谢通路。实时荧光定量PCR 验证结果与芯片杂交结果基本一致,印证了芯片杂交结果的有效性。上述发现提示,NO信号可能参与了WRKY转录因子介导的生物学调控功能,并为这些基因的进一步功能分析奠定基础。
中图分类号:
孟姣, 王海华, 向建华, 蒋丹, 彭喜旭, 贺欢欢. 水稻WRKY转录因子基因家族响应外源一氧化氮的表达谱分析[J]. 中国水稻科学, 2016, 30(2): 111-120.
Jiao MENG, Hai-hua WANG, Jian-hua XIANG, Dan JIANG, Xi-xu PENG, Huan-huan HE. Expression Profiles of Rice WRKY Transcription Factor Gene Family Responsive to Exogenous Nitric Oxide Application[J]. Chinese Journal OF Rice Science, 2016, 30(2): 111-120.
| 基因 Gene | 登录号 Accession No. | 正向引物 Forward primer (5'-3') | 反向引物 Reverse primer (5'-3') |
|---|---|---|---|
| OsWRKY15 | Os01g0656400 | GTCGTCGCCCTTCGTGTC | ATGAACTCCATGTCGTCTCCC |
| OsWRKY42 | Os02g0462800 | ATGGGAGCGTGTCCAACG | GCGGCCATCAGAAGCATAA |
| OsWRKY21 | Os01g0821600 | GCTGAGCAACAGAGCAGGTA | GCGTCAGTTATGCGTCAAG |
| OsWRKY77 | Os01g0584900 | CCTCGCTGCTGCTTCCTT | TTCAGGTCGCCTTGGTCC |
| OsWRKY71 | Os02g0181300 | AGGAATTGATGATTCGCTGTA | CGCCGATGATCGTTGGTT |
| OsWRKY24 | Os01g0826400 | TGCTCCTGACCTCCAGTATCTT | TTCCTCTGCTCGTCCTTGC |
| OsWRKY69 | Os08g0386200 | TGCTTCGACTTCGACCCG | TCTTCCCTCCTGCCACCA |
| Actin 1 | Os03g0718100 | ATCGCCCTGGACTATGAC | GAAACGCTCAGCACCAAT |
表1 候选基因的qRT-PCR引物
Table 1 Primers of the candidate genes for qRT-PCR.
| 基因 Gene | 登录号 Accession No. | 正向引物 Forward primer (5'-3') | 反向引物 Reverse primer (5'-3') |
|---|---|---|---|
| OsWRKY15 | Os01g0656400 | GTCGTCGCCCTTCGTGTC | ATGAACTCCATGTCGTCTCCC |
| OsWRKY42 | Os02g0462800 | ATGGGAGCGTGTCCAACG | GCGGCCATCAGAAGCATAA |
| OsWRKY21 | Os01g0821600 | GCTGAGCAACAGAGCAGGTA | GCGTCAGTTATGCGTCAAG |
| OsWRKY77 | Os01g0584900 | CCTCGCTGCTGCTTCCTT | TTCAGGTCGCCTTGGTCC |
| OsWRKY71 | Os02g0181300 | AGGAATTGATGATTCGCTGTA | CGCCGATGATCGTTGGTT |
| OsWRKY24 | Os01g0826400 | TGCTCCTGACCTCCAGTATCTT | TTCCTCTGCTCGTCCTTGC |
| OsWRKY69 | Os08g0386200 | TGCTTCGACTTCGACCCG | TCTTCCCTCCTGCCACCA |
| Actin 1 | Os03g0718100 | ATCGCCCTGGACTATGAC | GAAACGCTCAGCACCAAT |
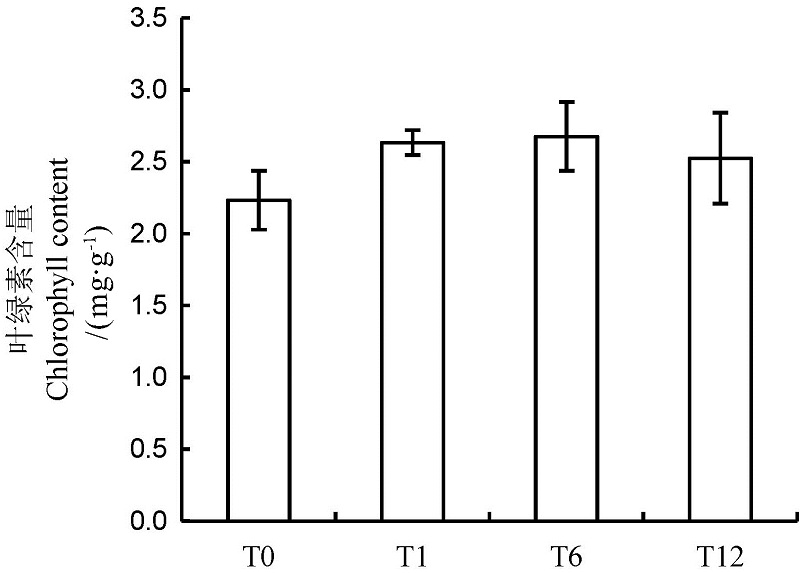
图1 NO处理不同时间后水稻叶片叶绿素含量 T0、T1、T6、T12分别表示NO处理0、1、6和12 h。下同。
Fig. 1. Chlorophyll contents in rice leaves after NO treatment for different time. NO treatments for 0,1,6 and 12 h are indicated as T0, T1, T6 and T12, respectively. The same as below.
| 基因 Gene | 探针 Probe | 表达倍数 Expression fold change | 组别 Group | ||
|---|---|---|---|---|---|
| T1/T0 | T6/T0 | T12/T0 | |||
| OsWRKY1 | Os01g0246700|mRNA|AY676928|CDS | 1.87 | 2.06 | 1.35 | Ⅱb |
| Os01g0246700|mRNA|AK105509|CDS+3'UTR | 2.72 | 1.54 | 2.19 | ||
| OsWRKY3 | Os03g0758000|mRNA|AY341859|CDS+3'UTR | 0.33 | 0.38 | 3.70 | Ⅰb |
| OsWRKY6 | Os03g0798500|COMBINER_EST|Os03g0798500|8 | 0.59 | 0.57 | 2.25 | Ⅱc |
| OsWRKY8 | Os05g0583000|mRNA|AK109568|CDS+3'UTR | 2.95 | 1.13 | 0.44 | Ⅱd |
| OsWRKY11 | Os01g0626400|mRNA|AY341856|CDS+3'UTR | 1.91 | 2.88 | 1.60 | Ⅰb |
| Os01g0626400|mRNA|AK108745|CDS+3'UTR | 2.23 | 2.57 | 1.63 | ||
| OsWRKY12 | Os01g0624700|mRNA|AK111416|CDS+3'UTR | 3.14 | 2.74 | 1.99 | Ⅱd |
| OsWRKY13 | Os01g0750100|mRNA|AK060018|CDS+3'UTR | 1.55 | 0.67 | 0.48 | Ⅱd |
| OsWRKY15 | Os01g0656400|mRNA|AY676926|CDS | 4.98 | 1.16 | 2.70 | Ⅲa |
| OsWRKY17 | Os01g0972800|mRNA|AY341842|CDS+3'UTR | 0.09 | 0.09 | 0.32 | Ⅰb |
| OsWRKY21 | Os01g0821600|mRNA|AK108657|CDS+3'UTR | 4.69 | 4.42 | 1.31 | Ⅲb |
| OsWRKY23 | Os01g0734000|mRNA|AK108909|CDS+3'UTR | 2.24 | 1.69 | 2.11 | Ⅰb |
| OsWRKY24 | Os01g0826400|mRNA|AK107199|CDS+3'UTR | 6.48 | 0.83 | 3.86 | Ⅰa |
| Os01g0826400|mRNA|AY341849|CDS+3'UTR | 5.39 | 0.63 | 3.33 | ||
| Os01g0826400|mRNA|AY676925|CDS | 5.92 | 0.75 | 3.65 | ||
| OsWRKY28 | Os06g0649000|mRNA|AK106282|CDS+3'UTR | 7.87 | 1.32 | 2.28 | Ⅱa |
| Os06g0649000|mRNA|AK119644|CDS+3'UTR | 6.77 | 1.05 | 1.91 | ||
| OsWRKY29 | Os07g0111400|mRNA|AY341858|CDS+3'UTR | 0.73 | 1.32 | 2.70 | Ⅰb |
| OsWRKY36 | Os04g0545000|mRNA|AK073695|CDS+3'UTR | 0.47 | 0.73 | 0.61 | Ⅰb |
| OsWRKY39 | Os02g0265200|mRNA|AK066775|CDS+3'UTR | 1.43 | 2.00 | 2.62 | Ⅱd |
| OsWRKY42 | Os02g0265200|mRNA|AK119593|CDS+3'UTR | 1.42 | 2.27 | 2.34 | Ⅱc |
| Os02g0462800|mRNA|AK110587|CDS+3'UTR | 5.51 | 3.80 | 2.01 | ||
| Os02g0462800|mRNA|AY676923|CDS | 5.01 | 3.14 | 2.05 | ||
| OsWRKY50 | Os11g0117600|COMBINER_EST|Os11g0117600|8 | 0.65 | 7.31 | 1.11 | Ⅲb |
| OsWRKY53 | Os05g0343400|mRNA|AY676929|CDS | 2.10 | 0.89 | 1.02 | Ⅰa |
| OsWRKY56 | Os12g0116600|mRNA|AK102093|CDS+3'UTR | 1.31 | 0.42 | 1.03 | Ⅲb |
| OsWRKY60 | Os03g0657400|COMBINER_EST|AU057193|6 | 1.01 | 0.58 | 0.44 | Ⅰb |
| OsWRKY62 | Os09g0417800|mRNA|AK067834|CDS+3'UTR | 1.48 | 1.65 | 2.03 | Ⅱa |
| OsWRKY67 | Os05g0183100|mRNA|AK066252|CDS+3'UTR | 1.13 | 0.66 | 0.28 | Ⅰb |
| OsWRKY69 | Os08g0386200|mRNA|AK111606|CDS+3'UTR | 3.05 | 0.98 | 2.77 | Ⅲa |
| OsWRKY71 | Os08g0386200|mRNA|AY676931|CDS | 3.33 | 0.88 | 2.80 | Ⅱa |
| Os02g0181300|mRNA|AK058773|CDS+3'UTR | 3.79 | 1.72 | 1.67 | ||
| Os02g0181300|mRNA|AY676927|CDS | 3.41 | 1.45 | 1.64 | ||
| OsWRKY72 | Os11g0490900|mRNA|AK108860|CDS+3'UTR | 2.32 | 0.89 | 1.75 | Ⅰb |
| OsWRKY77 | Os01g0584900|mRNA|AK108522|CDS+3'UTR | 1.10 | 1.80 | 2.31 | Ⅰb |
| Os01g0584900|mRNA|AY341846|CDS+3'UTR | 1.24 | 1.88 | 2.48 | ||
| OsWRKY79 | Os03g0335200|mRNA|AK105244|CDS+3'UTR | 0.66 | 1.83 | 2.53 | Ⅲb |
| OsWRKY88 | Os07g0596900|mRNA|AY341847|CDS+3'UTR | 0.94 | 1.53 | 2.13 | Ⅲa |
| OsWRKY104 | Os11g0117400|COMBINER_EST|CI444468|6 | 1.40 | 0.32 | 0.93 | Ⅲb |
| OsWRKY109 | Os05g0129800|COMBINER_EST|Os05g0129800|8 | 0.73 | 0.82 | 2.02 | Ⅱd |
| OsWRKY113 | Os06g0158100|mRNA|AY676932|CDS | 2.35 | 0.96 | 3.71 | Ⅲa |
表2 NO处理下水稻WRKY差异表达基因的芯片分析
Table 2 Microarray analysis of differentially expressed WRKY genes in rice under NO treatment.
| 基因 Gene | 探针 Probe | 表达倍数 Expression fold change | 组别 Group | ||
|---|---|---|---|---|---|
| T1/T0 | T6/T0 | T12/T0 | |||
| OsWRKY1 | Os01g0246700|mRNA|AY676928|CDS | 1.87 | 2.06 | 1.35 | Ⅱb |
| Os01g0246700|mRNA|AK105509|CDS+3'UTR | 2.72 | 1.54 | 2.19 | ||
| OsWRKY3 | Os03g0758000|mRNA|AY341859|CDS+3'UTR | 0.33 | 0.38 | 3.70 | Ⅰb |
| OsWRKY6 | Os03g0798500|COMBINER_EST|Os03g0798500|8 | 0.59 | 0.57 | 2.25 | Ⅱc |
| OsWRKY8 | Os05g0583000|mRNA|AK109568|CDS+3'UTR | 2.95 | 1.13 | 0.44 | Ⅱd |
| OsWRKY11 | Os01g0626400|mRNA|AY341856|CDS+3'UTR | 1.91 | 2.88 | 1.60 | Ⅰb |
| Os01g0626400|mRNA|AK108745|CDS+3'UTR | 2.23 | 2.57 | 1.63 | ||
| OsWRKY12 | Os01g0624700|mRNA|AK111416|CDS+3'UTR | 3.14 | 2.74 | 1.99 | Ⅱd |
| OsWRKY13 | Os01g0750100|mRNA|AK060018|CDS+3'UTR | 1.55 | 0.67 | 0.48 | Ⅱd |
| OsWRKY15 | Os01g0656400|mRNA|AY676926|CDS | 4.98 | 1.16 | 2.70 | Ⅲa |
| OsWRKY17 | Os01g0972800|mRNA|AY341842|CDS+3'UTR | 0.09 | 0.09 | 0.32 | Ⅰb |
| OsWRKY21 | Os01g0821600|mRNA|AK108657|CDS+3'UTR | 4.69 | 4.42 | 1.31 | Ⅲb |
| OsWRKY23 | Os01g0734000|mRNA|AK108909|CDS+3'UTR | 2.24 | 1.69 | 2.11 | Ⅰb |
| OsWRKY24 | Os01g0826400|mRNA|AK107199|CDS+3'UTR | 6.48 | 0.83 | 3.86 | Ⅰa |
| Os01g0826400|mRNA|AY341849|CDS+3'UTR | 5.39 | 0.63 | 3.33 | ||
| Os01g0826400|mRNA|AY676925|CDS | 5.92 | 0.75 | 3.65 | ||
| OsWRKY28 | Os06g0649000|mRNA|AK106282|CDS+3'UTR | 7.87 | 1.32 | 2.28 | Ⅱa |
| Os06g0649000|mRNA|AK119644|CDS+3'UTR | 6.77 | 1.05 | 1.91 | ||
| OsWRKY29 | Os07g0111400|mRNA|AY341858|CDS+3'UTR | 0.73 | 1.32 | 2.70 | Ⅰb |
| OsWRKY36 | Os04g0545000|mRNA|AK073695|CDS+3'UTR | 0.47 | 0.73 | 0.61 | Ⅰb |
| OsWRKY39 | Os02g0265200|mRNA|AK066775|CDS+3'UTR | 1.43 | 2.00 | 2.62 | Ⅱd |
| OsWRKY42 | Os02g0265200|mRNA|AK119593|CDS+3'UTR | 1.42 | 2.27 | 2.34 | Ⅱc |
| Os02g0462800|mRNA|AK110587|CDS+3'UTR | 5.51 | 3.80 | 2.01 | ||
| Os02g0462800|mRNA|AY676923|CDS | 5.01 | 3.14 | 2.05 | ||
| OsWRKY50 | Os11g0117600|COMBINER_EST|Os11g0117600|8 | 0.65 | 7.31 | 1.11 | Ⅲb |
| OsWRKY53 | Os05g0343400|mRNA|AY676929|CDS | 2.10 | 0.89 | 1.02 | Ⅰa |
| OsWRKY56 | Os12g0116600|mRNA|AK102093|CDS+3'UTR | 1.31 | 0.42 | 1.03 | Ⅲb |
| OsWRKY60 | Os03g0657400|COMBINER_EST|AU057193|6 | 1.01 | 0.58 | 0.44 | Ⅰb |
| OsWRKY62 | Os09g0417800|mRNA|AK067834|CDS+3'UTR | 1.48 | 1.65 | 2.03 | Ⅱa |
| OsWRKY67 | Os05g0183100|mRNA|AK066252|CDS+3'UTR | 1.13 | 0.66 | 0.28 | Ⅰb |
| OsWRKY69 | Os08g0386200|mRNA|AK111606|CDS+3'UTR | 3.05 | 0.98 | 2.77 | Ⅲa |
| OsWRKY71 | Os08g0386200|mRNA|AY676931|CDS | 3.33 | 0.88 | 2.80 | Ⅱa |
| Os02g0181300|mRNA|AK058773|CDS+3'UTR | 3.79 | 1.72 | 1.67 | ||
| Os02g0181300|mRNA|AY676927|CDS | 3.41 | 1.45 | 1.64 | ||
| OsWRKY72 | Os11g0490900|mRNA|AK108860|CDS+3'UTR | 2.32 | 0.89 | 1.75 | Ⅰb |
| OsWRKY77 | Os01g0584900|mRNA|AK108522|CDS+3'UTR | 1.10 | 1.80 | 2.31 | Ⅰb |
| Os01g0584900|mRNA|AY341846|CDS+3'UTR | 1.24 | 1.88 | 2.48 | ||
| OsWRKY79 | Os03g0335200|mRNA|AK105244|CDS+3'UTR | 0.66 | 1.83 | 2.53 | Ⅲb |
| OsWRKY88 | Os07g0596900|mRNA|AY341847|CDS+3'UTR | 0.94 | 1.53 | 2.13 | Ⅲa |
| OsWRKY104 | Os11g0117400|COMBINER_EST|CI444468|6 | 1.40 | 0.32 | 0.93 | Ⅲb |
| OsWRKY109 | Os05g0129800|COMBINER_EST|Os05g0129800|8 | 0.73 | 0.82 | 2.02 | Ⅱd |
| OsWRKY113 | Os06g0158100|mRNA|AY676932|CDS | 2.35 | 0.96 | 3.71 | Ⅲa |
| 组/亚组 Group/ subgroup | 差异表达基因数 Number of differentially expressed genes | 组/亚组的基因总数 Total number of genes in group/subgroup | 比例 Ratio /% |
|---|---|---|---|
| Ⅰ | 12 | 34 | 35.3 |
| Ⅱ | 11 | 30 | 36.7 |
| a | 3 | 4 | 75.0 |
| b | 1 | 8 | 12.5 |
| c | 2 | 7 | 28.6 |
| d | 5 | 11 | 45.6 |
| Ⅲ | 9 | 36 | 25.0 |
表3 NO处理下水稻WRKY差异表达基因在各组中的分布
Table 3 Group distribution of differentially expressed WRKY genes of rice responsive to nitric oxide.
| 组/亚组 Group/ subgroup | 差异表达基因数 Number of differentially expressed genes | 组/亚组的基因总数 Total number of genes in group/subgroup | 比例 Ratio /% |
|---|---|---|---|
| Ⅰ | 12 | 34 | 35.3 |
| Ⅱ | 11 | 30 | 36.7 |
| a | 3 | 4 | 75.0 |
| b | 1 | 8 | 12.5 |
| c | 2 | 7 | 28.6 |
| d | 5 | 11 | 45.6 |
| Ⅲ | 9 | 36 | 25.0 |
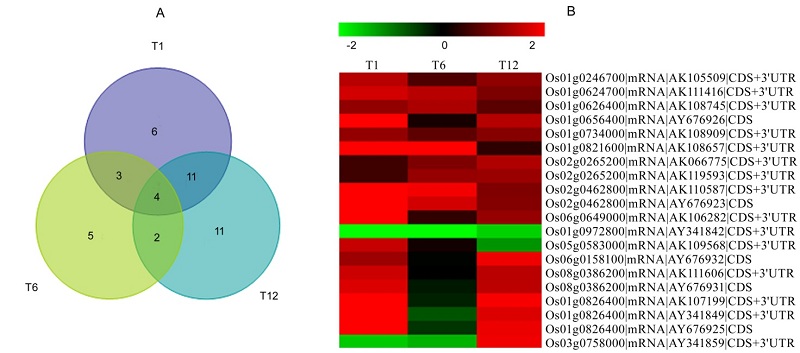
图3 NO响应的水稻WRKY基因的维恩图(A)和聚类热图(B) 比色刻度尺在上边。红色表示上调,绿色表示下调。
Fig.3. Venny analysis (A) and heatmap clustering (B) of NO-responsive rice WRKY gene. The color scale is shown on the top. Red, Up-regulated; Green, Down-regulated.
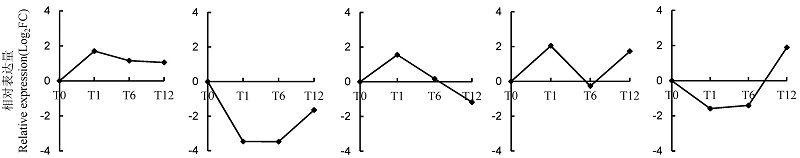
图5 NO响应的水稻WRKY基因的表达模式 Log2FC-Log2处理后的基因表达变化倍数。
Fig.5. Expression patterns of the NO-responsive rice WRKY genes. Log2FC indicates Log2 transformed expression fold change.
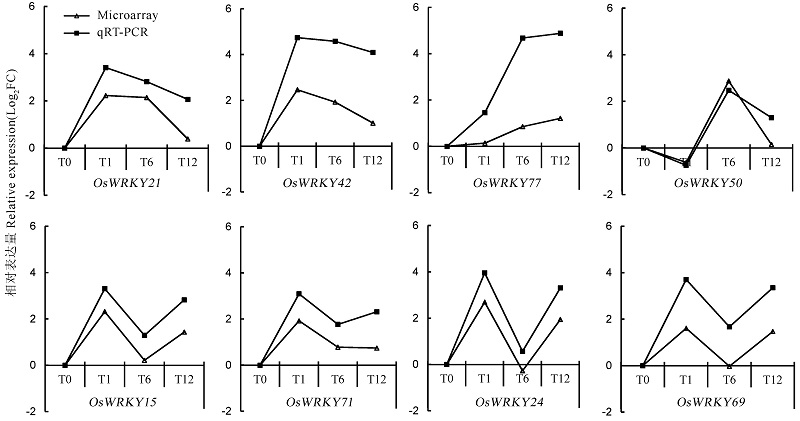
图6 NO响应的水稻WRKY基因表达水平的qRT-PCR验证 Log2FC-Log2处理后的基因表达变化倍数。
Fig. 6. Expression verification of NO-responsive rice WRKY genes by qRT-PCR. Log2FC indicates Log2 transformed expression fold change.
| [1] | Riaño-Pachón D M, Ruzicic S, Dreyer I, et al. PlnTFDB: An integrative plant transcription factor database.BMC Bioinform, 2007, 8: 42. |
| [2] | Eulgem T, Rushton P J, Robatzek S, et al.The WRKY superfamily of plant transcription factors.Trends Plant Sci, 2000, 5: 199-206. |
| [3] | Pandey S P, Somssich I E.The role of WRKY transcription factors in plant immunity.Plant Physiol, 2009, 150: 1648-1655. |
| [4] | Agarwal P, Reddy M P, Chikara J.WRKY: Its structure, evolutionary relationship, DNA-binding selectivity, role in stress tolerance and development of plants.Mol Biol Rep, 2011, 38: 3883-3896. |
| [5] | Chen L, Song Y, Li S, et al.The role of WRKY transcription factors in plant abiotic stresses.Biochim Biophys Acta, 2012, 1819: 120-128. |
| [6] | Li J, Brader G, Palva E T.The WRKY70 transcription factor: A node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell, 2004, 16: 319-331. |
| [7] | Qiu D, Xiao J, Ding X, et al.OsWRKY13 mediates rice disease resistance by regulating defense-related genes in salicylate- and jasmonate-dependent signaling.Mol Plant Microbe Interact, 2007, 20: 492-499. |
| [8] | Wang H, Meng J, Peng X, et al.Rice WRKY4 acts as a transcriptional activator mediating defense responses toward Rhizoctonia solani, the causing agent of rice sheath blight.Plant Mol Biol, 2005, 89(1/2): 157-171. |
| [9] | Shang Y, Yan L, Liu Z Q, et al.The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition.Plant Cell, 2010, 22, 1909-1935. |
| [10] | Ryu H S, Han M, Lee S K, et al.A comprehensive expression analysis of the WRKY gene superfamily in rice plants during defense response.Plant Cell Rep, 2006, 25: 836-847. |
| [11] | Ramamoorthy R, Jiang S Y, Kumar N, et al.A comprehensive transcriptional profiling of the WRKY gene family in rice under various abiotic and phytohormone treatments.Plant Cell Physiol, 2008, 49: 865-879. |
| [12] | Lamotte O, Courtois C, Pugin L B A, et al. Nitric oxide in plants: The biosynthesis and cell signalling properties of a fascinating molecule.Planta, 2005, 221: 1-4. |
| [13] | Gould K, Lamotte O, Klinguer A, et al.Nitric oxide production in tobacco leaf cells: A generalized stress response?Plant Cell Environ, 2003, 26: 1851-1862. |
| [14] | Delledonne M.NO news is good news for plants.Curr Opin Plant Biol, 2005, 8: 390-396. |
| [15] | Palmieri M C, Sell S, Huang X, et al.Nitric oxide-responsive genes and promoters in Arabidopsis thaliana: A bioinformatics approach.J Exp Bot, 2008, 59:177-186. |
| [16] | Mukhtar M S, Deslandes L, Auriac M-C, et al.The Arabidopsis transcription factor WRKY27 influences wilt disease symptom development caused by Ralstonia solanacearum.Plant J, 2008, 56: 935-947. |
| [17] | Arnon D I.Copper enzymes in isolated chloroplasts: Polyphenoloxidase in Beta vulgaris. Plant Physiol, 1949, 24:1-15. |
| [18] | Rice WRKY Working Group. Nomenclature report on rice WRKY's-Conflict regarding gene names and its solution.Rice, 2012, 5: 3. |
| [19] | Wu K L, Guo Z J, Wang H H, et al.The WRKY family of transcription factors in rice and Arabidopsis and their origins.DNA Res, 2005, 12: 9-26. |
| [20] | Dong J, Chen C, Chen Z.Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response.Plant Mol Biol, 2003, 51: 21-37. |
| [21] | Kalde M, Barth M, Somssich I E, et al.Members of the Arabidopsis WRKY group Ⅲ transcription factors are part of different plant defense signaling pathways.Mol Plant Microbe Interact, 2003, 16: 295-305. |
| [22] | Chujo T, Miyamoto K, Shimogawa T, et al.OsWRKY28, a PAMP-responsive transrepressor, negatively regulates innate immune responses in rice against rice blast fungus.Plant Mol Biol, 2013, 82: 23-37. |
| [23] | Peng Y, Bartley L E, Chen X, et al.OsWRKY62 is a negative regulator of basal and Xa21-mediated defense against Xanthomonas oryzae pv. oryzae in rice.Mol Plant, 2008, 1: 446-458. |
| [24] | Yokotani N, Sato Y, Tanabe S, et al.WRKY76 is a rice transcriptional repressor playing opposite roles in blast disease resistance and cold stress tolerance.J Exp Bot, 2013, 64(16): 5085-5097. |
| [25] | Liu X, Bai X, Wang X, et al.OsWRKY71, a rice transcription factor, is involved in rice defense response.J Plant Physiol, 2007, 164: 969-979. |
| [26] | Park C Y, Lee J H, Yoo J H, et al.WRKY group IId transcription factors interact with calmodulin.FEBS Lett, 2005, 579: 1545-1550. |
| [27] | Durner J, Wendehenne D, Klessig D F.Defense gene induction in tobacco by nitric oxide, cyclic GMP, and cyclic ADP-ribose.Proc Natl Acad Sci USA, 1998, 95: 10328-10333. |
| [28] | Liu X Q, Bai X Q, Qian Q, et al.OsWRKY03, a rice transcriptional activator that functions in defense signaling pathway upstream of OsNPR1.Cell Res, 2005, 15(8): 593-603. |
| [29] | Jing S, Zhou X, Song Y, et al.Heterologous expression of OsWRKY23 gene enhances pathogen defense and dark-induced leaf senescence in Arabidopsis. Plant Growth Regul, 2009, 58:181-190. |
| [30] | Song Y, Jing S, Yu D.Overexpression of the stress-induced OsWRKY08 improves osmotic stress tolerance inArabidopsis. Chin Sci Bull, 2009, 54: 4671-4678. |
| [31] | Cheng H, Liu H, Deng Y, et al.The WRKY45-2 WRKY13 WRKY42 transcriptional regulatory cascade is required for rice resistance to fungal pathogen.Plant Physiol, 2015, 167(3): 1087-1099. |
| [32] | Zhang Z L, Shin M, Zou X, et al.A negative regulator encoded by a rice WRKY gene represses both abscisic acid and gibberellins signaling in aleurone cells.Plant Mol Biol, 2009, 70: 139-151. |
| [33] | Sperotto R A, Boff T, Duarte G L, et al.Increased senescence-associated gene expression and lipid peroxidation induced by iron deficiency in rice roots.Plant Cell Rep, 2008, 27: 183-195. |
| [34] | 黄盼盼. 18个水稻OsWRKY基因的克隆和OsWRKY21功能分析. 广州: 中山大学, 2010. |
| Huang P P.Cloning 18 OsWRKY genes and functional analysis of OsWRKY21. GuangZhou:Sun Yat-sen University, 2010. (in Chinese with English abstract) |
| [1] | 郭展, 张运波. 水稻对干旱胁迫的生理生化响应及分子调控研究进展[J]. 中国水稻科学, 2024, 38(4): 335-349. |
| [2] | 韦还和, 马唯一, 左博源, 汪璐璐, 朱旺, 耿孝宇, 张翔, 孟天瑶, 陈英龙, 高平磊, 许轲, 霍中洋, 戴其根. 盐、干旱及其复合胁迫对水稻产量和品质形成影响的研究进展[J]. 中国水稻科学, 2024, 38(4): 350-363. |
| [3] | 许丹洁, 林巧霞, 李正康, 庄小倩, 凌宇, 赖美玲, 陈晓婷, 鲁国东. OsOPR10正调控水稻对稻瘟病和白叶枯病的抗性[J]. 中国水稻科学, 2024, 38(4): 364-374. |
| [4] | 候小琴, 王莹, 余贝, 符卫蒙, 奉保华, 沈煜潮, 谢杭军, 王焕然, 许用强, 武志海, 王建军, 陶龙兴, 符冠富. 黄腐酸钾提高水稻秧苗耐盐性的作用途径分析[J]. 中国水稻科学, 2024, 38(4): 409-421. |
| [5] | 胡继杰, 胡志华, 张均华, 曹小闯, 金千瑜, 章志远, 朱练峰. 根际饱和溶解氧对水稻分蘖期光合及生长特性的影响[J]. 中国水稻科学, 2024, 38(4): 437-446. |
| [6] | 刘福祥, 甄浩洋, 彭焕, 郑刘春, 彭德良, 文艳华. 广东省水稻孢囊线虫病调查与鉴定[J]. 中国水稻科学, 2024, 38(4): 456-461. |
| [7] | 陈浩田, 秦缘, 钟笑涵, 林晨语, 秦竞航, 杨建昌, 张伟杨. 水稻根系和土壤性状与稻田甲烷排放关系的研究进展[J]. 中国水稻科学, 2024, 38(3): 233-245. |
| [8] | 缪军, 冉金晖, 徐梦彬, 卜柳冰, 王平, 梁国华, 周勇. 过量表达异三聚体G蛋白γ亚基基因RGG2提高水稻抗旱性[J]. 中国水稻科学, 2024, 38(3): 246-255. |
| [9] | 尹潇潇, 张芷菡, 颜绣莲, 廖蓉, 杨思葭, 郭岱铭, 樊晶, 赵志学, 王文明. 多个稻曲病菌效应因子的信号肽验证和表达分析[J]. 中国水稻科学, 2024, 38(3): 256-265. |
| [10] | 朱裕敬, 桂金鑫, 龚成云, 罗新阳, 石居斌, 张海清, 贺记外. 全基因组关联分析定位水稻分蘖角度QTL[J]. 中国水稻科学, 2024, 38(3): 266-276. |
| [11] | 魏倩倩, 汪玉磊, 孔海民, 徐青山, 颜玉莲, 潘林, 迟春欣, 孔亚丽, 田文昊, 朱练峰, 曹小闯, 张均华, 朱春权. 信号分子硫化氢参与硫肥缓解铝对水稻生长抑制作用的机制[J]. 中国水稻科学, 2024, 38(3): 290-302. |
| [12] | 周甜, 吴少华, 康建宏, 吴宏亮, 杨生龙, 王星强, 李昱, 黄玉峰. 不同种植模式对水稻籽粒淀粉含量及淀粉关键酶活性的影响[J]. 中国水稻科学, 2024, 38(3): 303-315. |
| [13] | 关雅琪, 鄂志国, 王磊, 申红芳. 影响中国水稻生产环节外包发展因素的实证研究:基于群体效应视角[J]. 中国水稻科学, 2024, 38(3): 324-334. |
| [14] | 许用强, 姜宁, 奉保华, 肖晶晶, 陶龙兴, 符冠富. 水稻开花期高温热害响应机理及其调控技术研究进展[J]. 中国水稻科学, 2024, 38(2): 111-126. |
| [15] | 吕海涛, 李建忠, 鲁艳辉, 徐红星, 郑许松, 吕仲贤. 稻田福寿螺的发生、危害及其防控技术研究进展[J]. 中国水稻科学, 2024, 38(2): 127-139. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||