
中国水稻科学 ›› 2017, Vol. 31 ›› Issue (3): 223-231.DOI: 10.16819/j.1001-7216.2017.7029 223
• • 下一篇
沈兰1,2,#, 李健3,#, 付亚萍2, 王俊杰2, 华宇峰2, 焦晓真2, 严长杰1,*( ), 王克剑2,*(
), 王克剑2,*( )
)
收稿日期:2017-03-09
修回日期:2017-03-20
出版日期:2017-05-10
发布日期:2017-05-10
通讯作者:
严长杰,王克剑
基金资助:
Lan SHEN1,2,#, Jian LI3,#, Yaping FU2, Junjie WANG2, Yufeng HUA2, Xiaozhen JIAO2, Changjie YAN1,*( ), Kejian WANG2,*(
), Kejian WANG2,*( )
)
Received:2017-03-09
Revised:2017-03-20
Online:2017-05-10
Published:2017-05-10
Contact:
Changjie YAN, Kejian WANG
摘要:
【目的】基因组定点编辑技术已成为分子育种的重要手段。本研究拟对GS3和Gn1a功能缺失突变对目标性状的改良效应进行分析,以期为培育高产水稻提供理论基础。【方法】利用CRISPR/Cas9系统,以控制粒型基因GS3和控制每穗粒数基因Gn1a为编辑对象,构建了共敲除载体pC1300-2×35S::Cas9-gGS3-gGn1a,用农杆菌介导法转化4个优质水稻品种,分析了基因突变的特征和相应农艺性状。【结果】构建的敲除载体成功地实现了对GS3和Gn1a基因的定点编辑。在4个转化受体的T0代均分别获得了gs3和gs3gn1a的移码突变体。对T1代中无选择标记突变体的农艺性状分析表明,突变体 gs3和gs3gn1a与野生型相比粒长变长,千粒重增加;突变体gs3gn1a与突变体 gs3相比,每穗粒数显著增加。【结论】利用CRISPR/Cas9系统进行水稻基因编辑可以快速改良品种的目标性状,在水稻品种的定向改良方面具有巨大的潜力。
中图分类号:
沈兰, 李健, 付亚萍, 王俊杰, 华宇峰, 焦晓真, 严长杰, 王克剑. 利用CRISPR/Cas9系统定向改良水稻粒长和穗粒数性状[J]. 中国水稻科学, 2017, 31(3): 223-231.
Lan SHEN, Jian LI, Yaping FU, Junjie WANG, Yufeng HUA, Xiaozhen JIAO, Changjie YAN, Kejian WANG. Orientation Improvement of Grain Length and Grain Number in Rice by Using CRISPR/Cas9 System[J]. Chinese Journal OF Rice Science, 2017, 31(3): 223-231.
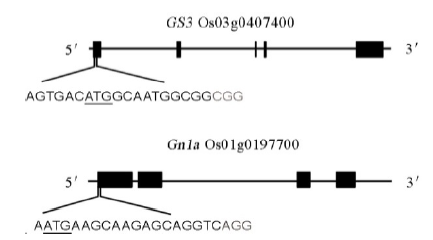
图1 GS3和Gn1a靶点位置带下划线的字母为起始密码子,灰色字母为PAM序列。
Fig. 1. Schematic diagram of the targeted sites in GS3 and Gn1a. The underlined letters are the initiation codons. The gray letters are the protospacer adjacent motif (PAM) sequences.

图2 CRISPR/Cas9载体构建流程中间载体SK-gRNA包含U3启动子和sgRNA骨架结构,双元载体pC1300-Cas9包含2×35S启动子和Cas9蛋白。GS3, Gn1a 的gRNA 分别用KpnⅠ/ SalⅠ和 XhoⅠ/ BglⅡ酶切,并通过一步法连入用KpnⅠ和 BamHⅠ酶切的pC1300-Cas9载体中。
Fig. 2. Flow diagram of CRISPR/Cas9 system for construction. The intermediate vector SK-gRNA contains the U3 promotor and sgRNA scaffold. Binary vector pC1300-Cas9 contains the 2×35S promotor and a Cas9 protein. Two gRNA scaffolds with GS3, Gn1a gene targets are respectively digested with KpnⅠ/ SalⅠ, and XhoⅠ/ BglⅡ, and cloned into pC1300-Cas9 between the KpnⅠand BamHⅠsites in a one-step ligation.
| 引物名称 Primer name | 引物序列 Sequence(5′-3′) | |
|---|---|---|
| GS3-g++ | GGCAGTGACATGGCAATGGCGG | |
| GS3-g–– | AAACCCGCCATTGCCATGTCAC | |
| Gn1a-g++ | GGCAATGAAGCAAGAGCAGGTC | |
| Gn1a-g–– | AAACGACCTGCTCTTGCTTCAT | |
| GS3-JC-F | CTATACATAGCTGCTGCAC | |
| GS3-JC-R | GACAGATAGCAAGCCGTAC | |
| Gn1a-JC-F | TTCCATCGTCAGCACACAAA | |
| Gn1a-JC-R | ACGGAGAGGTTGCCAAAGTC | |
| Hyg-F1 | GCTGTTATGCGGCCATTGTC | |
| Hyg-R1 | GACGTCTGTCGAGAAGTTTC | |
| Cas9-F2 | ACCAGACACGAGACGACTAA | |
| pC1300-R2 | ATCGGTGCGGGCCTCTTC | |
| Actin-F | TGCTATGTACGTCGCCATCCA | |
| Actin-R | AATGAGTAACCACGCTCCGTC | |
| Gn1a-F | CCATGGTATGCATGCAACACCATG | |
| Gn1a-R | CGTTGTCACGTACTCCCTCCGTA | |
| GS3-F | CTATACATAGCTGCTGCACCGTCT | |
| GS3-R | CAATCACGTACTCATCATGGCAGCA | |
| T3 | ATTAACCCTCACTAAAGGGA | |
表1 本研究所用的引物
Table 1 Primers used in this research.
| 引物名称 Primer name | 引物序列 Sequence(5′-3′) | |
|---|---|---|
| GS3-g++ | GGCAGTGACATGGCAATGGCGG | |
| GS3-g–– | AAACCCGCCATTGCCATGTCAC | |
| Gn1a-g++ | GGCAATGAAGCAAGAGCAGGTC | |
| Gn1a-g–– | AAACGACCTGCTCTTGCTTCAT | |
| GS3-JC-F | CTATACATAGCTGCTGCAC | |
| GS3-JC-R | GACAGATAGCAAGCCGTAC | |
| Gn1a-JC-F | TTCCATCGTCAGCACACAAA | |
| Gn1a-JC-R | ACGGAGAGGTTGCCAAAGTC | |
| Hyg-F1 | GCTGTTATGCGGCCATTGTC | |
| Hyg-R1 | GACGTCTGTCGAGAAGTTTC | |
| Cas9-F2 | ACCAGACACGAGACGACTAA | |
| pC1300-R2 | ATCGGTGCGGGCCTCTTC | |
| Actin-F | TGCTATGTACGTCGCCATCCA | |
| Actin-R | AATGAGTAACCACGCTCCGTC | |
| Gn1a-F | CCATGGTATGCATGCAACACCATG | |
| Gn1a-R | CGTTGTCACGTACTCCCTCCGTA | |
| GS3-F | CTATACATAGCTGCTGCACCGTCT | |
| GS3-R | CAATCACGTACTCATCATGGCAGCA | |
| T3 | ATTAACCCTCACTAAAGGGA | |
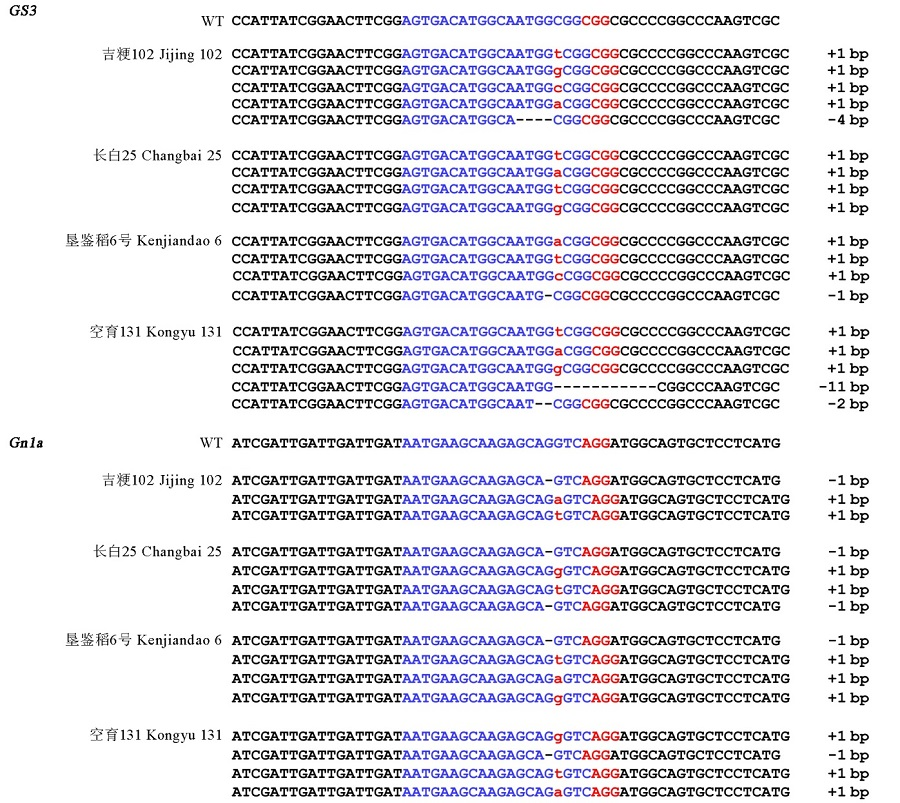
图3 4个水稻品种T0代GS3和Gn1a 突变类型分析靶序列用蓝色标注,PAM序列用红色标注,碱基插入的用红色小写字母表示,缺失碱基用黑色连字符表示。
Fig. 3. Mutation types at the GS3 and Gn1a loci of the four varieties in T0 generation. The targeted sequence is highlighted in blue and the PAM sequence in red. Mutations with 1 bp insertion are represented by red lowercase letters. The deleted sequences are shown by black hyphens.
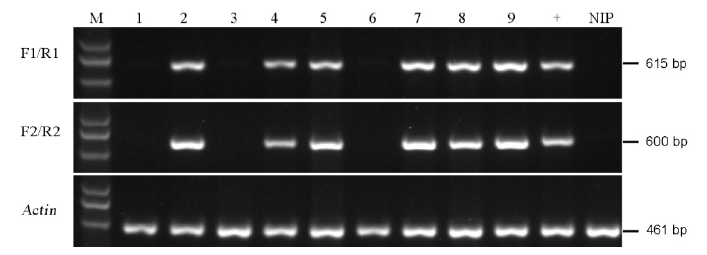
图4 PCR鉴定无选择标记基因的突变株 M–标记D5000;1~9–T1代转基因株系;+,转基因阳性对照;NIP–无选择标记基因的对照(日本晴)。
Fig. 4. PCR identification of the marker-free transgenic plants. M, Marker D5000; Lanes 1 to 9, T1 transgenic lines; +, Positive control of transgenic line, NIP, Negative control(Nipponbare).

图5 4个品种T1代株型 J102–吉粳102; Ch 25–长白25; Ken 6–垦鉴稻6号; Ko 131–空育131。
Fig. 5. Grass morphology of the plants of the four varieties in T1 generation. J102, Jijing 102; Ch 25, Changbai 25; Ken 6, Kenjiandao 6; Ko 131, Kongyu 131.

图6 T1代4个水稻品种及其突变体粒型 A, D, G, J分别为吉粳102、长白25、垦鉴稻6号和空育131水稻品种及其突变体粒型;B, E, H, K 和C, F, I, L分别为其对应的粒长和粒宽。*, **分别表示在0.05和0.01水平上显著差异。图7~8同。
Fig. 6. Grain size of the four varieties and their mutants in T1 generation. A, D, G, J, Grain shapes of Jijing 102, Changbai 25, Kenjiandao 6 and Kongyu 131 and their mutants, respectively. B, E, H, K and C, F, I, L show the grain length and grain width, respectively. *, **, Significant difference at 0.05 and 0.01 levels, respectively. The same as in Fig 7 and 8.
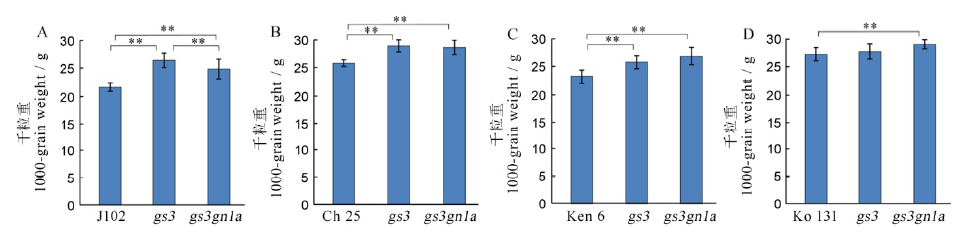
图7 T1代4个水稻品种及其突变体的千粒重 A, B, C, D分别为吉粳102、长白25、垦鉴稻6号和空育131水稻品种及其突变体的千粒重。
Fig. 7. 1000-grain weight among the four varieties and their mutants in T1 generation. A, B, C, D shows the 1000-grain weight of Jijing 102, Changbai 25, Kenjiandao 6 and Kongyu 131 and their mutants, respectively.
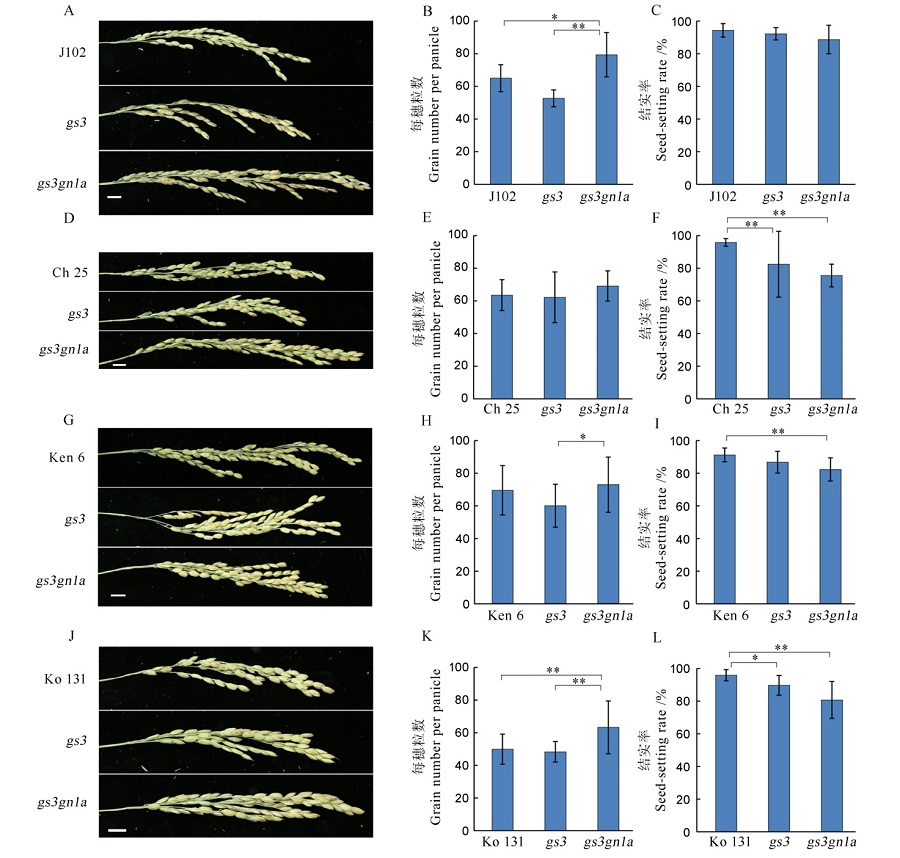
图8 T1代4个水稻品种及其突变体的每穗粒数和结实率 A, D, G, J分别为吉粳102、长白25、垦鉴稻6号和空育131水稻品种及其突变体的穗型,B, E, H, K和C, F, I, L分别为其对应的每穗粒数和结实率。
Fig. 8. Grain number per panicle and the seed-setting rate among the four varieties in T1 generation and their mutants. A, D, G, J indicate the panicle phenotype of Jijing 102, Changbai 25, Kenjiandao 6 and Kongyu 131 and their mutants, respectively. B, E, H, K and C, F, I, L show the grain number per panicle and the seed-setting rate, respectively.
| [1] | Wiedenheft B, Sternberg S H, Doudna J A.RNA-guided genetic silencing systems in bacteria and archaea.Nature, 2012, 482: 331-338. |
| [2] | Cong L, Ran F A, Cox D, Lin S, Barretto R, Habib N, Hsu P D, Wu X, Jiang W, Marraffini L A, Zhang F.Multiplex genome engineering using CRISPR/Cas systems. Science, 2013, 339: 819-823. |
| [3] | Shan Q, Wang Y, Li J, Zhang Y, Chen K, Liang Z, Zhang K, Liu J, Xi J J, Qiu J L, Gao C.Targeted genome modification of crop plants using a CRISPR/Cas system. Nat Biotechnol, 2013, 31: 686-688. |
| [4] | DiCarlo J E, Norville J E, Mali P, Rios X, Aach J, Church G M. Genome engineering in Saccharomyces cerevisiae using CRISPR/Cas systems.Nucleic Acids Res, 2013, 41: 4336-4343. |
| [5] | Jakociunas T, Bonde I, Herrgard M, Harrison SJ, Kristensen M, Pedersen L E, Jensen M K, Keasling J D.Multiplex metabolic pathway engineering using CRISPR/Cas9 in Saccharomyces cerevisiae.Metab Eng, 2015, 28: 213-222. |
| [6] | Zhou X, Xin J, Fan N, Zou Q, Huang J, Ouyang Z, Zhao Y, Zhao B, Liu Z, Lai S, Yi X, Guo L, Esteban M A, Zeng Y, Yang H, Lai L.Generation of CRISPR/Cas 9-mediated gene-targeted pigs via somatic cell nuclear transfer.Cell Mol Life Sci, 2015, 72: 1175-1184. |
| [7] | Zou Q, Wang X, Liu Y, Ouyang Z, Long H, Wei S, Xin J, Zhao B, Lai S, Shen J, Ni Q, Yang H, Zhong H, Li L, Hu M, Zhang Q, Zhou Z, He J, Yan Q, Fan N, Zhao Y, Liu Z, Guo L, Huang J, Zhang G, Ying J, Lai L, Gao X.Generation of gene-target dogs using CRISPR/Cas 9 system.J Mol Cell Biol, 2015, 7: 580-583. |
| [8] | Wang H, Yang H, Shivalila C S, Dawlaty M M, Cheng A W, Zhang F, Jaenisch R.One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas- mediated genome engineering.Cell, 2013, 153: 910-918. |
| [9] | Hoshijima K, Jurynec M J, Grunwald D J.Precise editing of the zebrafish genome made simple and efficient.Dev Cell, 2016, 36: 654-667. |
| [10] | Gratz S J, Harrison M M, Wildonger J, O'Connor-Giles K M. Precise genome editing of drosophila with CRISPR/Cas RNA-guided Cas9.Methods Mol Biol, 2015, 1311: 335-348. |
| [11] | Mao Y, Zhang H, Xu N, Zhang B, Gou F, Zhu J K.Application of the CRISPR-Cas system for efficient genome engineering in plants. Mol Plant, 2013, 6: 2008-2011. |
| [12] | Puchta H.Applying crispr/cas for genome engineering in plants: The best is yet to come.Curr Opin Plant Biol, 2016, 36: 1-8. |
| [13] | Jiang W, Zhou H, Bi H, Fromm M, Yang B, Weeks D P.Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice.Nucleic Acids Res, 2013, 41: e188. |
| [14] | Feng Z, Zhang B, Ding W, Liu X, Yang D L, Wei P, Cao F, Zhu S, Zhang F, Mao Y, Zhu J K.Efficient genome editing in plants using a CRISPR/Cas system.Cell Res, 2013, 23: 1229-1232. |
| [15] | Wang C, Shen L, Fu Y, Yan C, Wang K.A simple crispr/cas9 system for multiplex genome editing in rice. J Genet Genom, 2015, 42: 703-706. |
| [16] | Zhang Z J, Mao Y F, Ha S, Liu W S, Botella J R, Zhu J K.A multiplex CRISPR/Cas9 platform for fast and efficient editing of multiple genes in Arabidopsis.Plant Cell Rep, 2016, 35: 1519-1533. |
| [17] | Ma X L, Zhang Q Y, Zhu Q L, Liu W, Chen Y, Qiu R, Wang B, Yang Z F, Li H Y, Lin Y R, Xie Y Y, Shen R X, Chen S F, Wang Z, Chen Y L, Guo J X, Chen L T, Zhao X C, Dong Z C, Liu Y G.A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants.Mol Plant, 2015, 8: 1274-1284. |
| [18] | Ma X, Chen L, Zhu Q, Chen Y, Liu Y G.Rapid decoding of sequence-specific nuclease-induced heterozygous and biallelic mutations by direct sequencing of PCR products.Mol Plant, 2015, 8: 1285-1287. |
| [19] | Xu R F, Li H, Qin R Y, Li J, Qiu C H, Yang Y C, Ma H, Li L, Wei P C, Yang J B.Generation of inheritable and "transgene clean" targeted genome-modified rice in later generations using the CRISPR/Cas9 system.Sci Rep, 2015, 5: 11491 |
| [20] | Li J, Sun Y, Du J, Zhao Y, Xia L.Generation of targeted point mutations in rice by a modified CRISPR/CAS9 system. Mol Plant, 2017, 10(3): 526-529. |
| [21] | Lu Y, Zhu J K.Precise editing of a target base in the rice genome using a modified CRISPR/Cas9 system.Mol Plant, 2017, 10(3): 523-525. |
| [22] | Fan C, Xing Y, Mao H, Lu T, Han B, Xu C, Li X, Zhang Q.Gs3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor Appl Genet, 2006, 112: 1164-1171. |
| [23] | Mao H, Sun S, Yao J, Wang C, Yu S, Xu C, Li X, Zhang Q.Linking differential domain functions of theGS3 protein to natural variation of grain size in rice.Proc Natl Acad Sci U S A, 2010, 107: 19579-19584. |
| [24] | Ashikari M, Sakakibara H, Lin S, Yamamoto T, Takashi T, Nishimura A, Angeles E R, Qian Q, Kitano H, Matsuoka M.Cytokinin oxidase regulates rice grain production.Science, 2005, 309: 741-745. |
| [25] | Shen L, Wang C, Fu Y, Wang J, Liu Q, Zhang X, Yan C, Qian Q, Wang K.QTL editing confers opposing yield performance in different rice varieties.J Integr Plant Biol, 2016, DOI: 10.1111/jipb.12501 |
| [26] | Hiei Y, Ohta S, Komari T, Kumashiro T.Efficient transformation of rice(Oryza sativa L.) mediated by agrobacterium and sequence analysis of the boundaries of the t-DNA.Plant J, 1994, 6: 271-282. |
| [27] | Li M, Li X, Zhou Z, Wu P, Fang M, Pan X, Lin Q, Luo W, Wu G, Li H. Reassessment of the four yield-related genes Gn1a, DEP1, GS3, and IPA1 in rice using a CRISPR/Cas9 system. Front Plant Sci, 2016, 7: |
| [28] | Feng C, Yuan J, Wang R, Liu Y, Birchler J A, Han F.Efficient targeted genome modification in maize using CRISPR/Cas 9 system. J Genet Genom, 2016, 43: 37-43. |
| [29] | Svitashev S, Young J K, Schwartz C, Gao H, Falco S C, Cigan A M.Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using Cas9 and guide RNA.Plant Physiol, 2015, 169: 931-945 |
| [30] | Shan Q, Wang Y, Li J, Gao C.Genome editing in rice and wheat using the CRISPR/Cas system.Nat Protoc, 2014, 9: 2395-2410. |
| [31] | Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C, Qiu J L.Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew.Nat Biotechnol, 2014, 32: 947-951. |
| [32] | Lawrenson T, Shorinola O, Stacey N, Li C, Ostergaard L, Patron N, Uauy C, Harwood W.Induction of targeted, heritable mutations in barley and Brassica oleracea using RNA-guidedCas9 nuclease. Genome Biol, 2015, 16: 258. |
| [33] | Cai Y, Chen L, Liu X, Sun S, Wu C, Jiang B, Han T, Hou W.CRISPR/Cas 9-mediated genome editing in soybean hairy roots.PLoS One, 2015, 10: e0136064. |
| [34] | Jacobs T B, LaFayette P R, Schmitz R J, Parrott W A. Targeted genome modifications in soybean with CRISPR/Cas 9.BMC Biotechnol, 2015, 15: 16. |
| [35] | Xing Y, Zhang Q.Genetic and molecular bases of rice yield.Annu Rev Plant Biol, 2010, 61: 421-442. |
| [1] | 郭展, 张运波. 水稻对干旱胁迫的生理生化响应及分子调控研究进展[J]. 中国水稻科学, 2024, 38(4): 335-349. |
| [2] | 韦还和, 马唯一, 左博源, 汪璐璐, 朱旺, 耿孝宇, 张翔, 孟天瑶, 陈英龙, 高平磊, 许轲, 霍中洋, 戴其根. 盐、干旱及其复合胁迫对水稻产量和品质形成影响的研究进展[J]. 中国水稻科学, 2024, 38(4): 350-363. |
| [3] | 许丹洁, 林巧霞, 李正康, 庄小倩, 凌宇, 赖美玲, 陈晓婷, 鲁国东. OsOPR10正调控水稻对稻瘟病和白叶枯病的抗性[J]. 中国水稻科学, 2024, 38(4): 364-374. |
| [4] | 候小琴, 王莹, 余贝, 符卫蒙, 奉保华, 沈煜潮, 谢杭军, 王焕然, 许用强, 武志海, 王建军, 陶龙兴, 符冠富. 黄腐酸钾提高水稻秧苗耐盐性的作用途径分析[J]. 中国水稻科学, 2024, 38(4): 409-421. |
| [5] | 胡继杰, 胡志华, 张均华, 曹小闯, 金千瑜, 章志远, 朱练峰. 根际饱和溶解氧对水稻分蘖期光合及生长特性的影响[J]. 中国水稻科学, 2024, 38(4): 437-446. |
| [6] | 刘福祥, 甄浩洋, 彭焕, 郑刘春, 彭德良, 文艳华. 广东省水稻孢囊线虫病调查与鉴定[J]. 中国水稻科学, 2024, 38(4): 456-461. |
| [7] | 陈浩田, 秦缘, 钟笑涵, 林晨语, 秦竞航, 杨建昌, 张伟杨. 水稻根系和土壤性状与稻田甲烷排放关系的研究进展[J]. 中国水稻科学, 2024, 38(3): 233-245. |
| [8] | 缪军, 冉金晖, 徐梦彬, 卜柳冰, 王平, 梁国华, 周勇. 过量表达异三聚体G蛋白γ亚基基因RGG2提高水稻抗旱性[J]. 中国水稻科学, 2024, 38(3): 246-255. |
| [9] | 尹潇潇, 张芷菡, 颜绣莲, 廖蓉, 杨思葭, 郭岱铭, 樊晶, 赵志学, 王文明. 多个稻曲病菌效应因子的信号肽验证和表达分析[J]. 中国水稻科学, 2024, 38(3): 256-265. |
| [10] | 朱裕敬, 桂金鑫, 龚成云, 罗新阳, 石居斌, 张海清, 贺记外. 全基因组关联分析定位水稻分蘖角度QTL[J]. 中国水稻科学, 2024, 38(3): 266-276. |
| [11] | 魏倩倩, 汪玉磊, 孔海民, 徐青山, 颜玉莲, 潘林, 迟春欣, 孔亚丽, 田文昊, 朱练峰, 曹小闯, 张均华, 朱春权. 信号分子硫化氢参与硫肥缓解铝对水稻生长抑制作用的机制[J]. 中国水稻科学, 2024, 38(3): 290-302. |
| [12] | 周甜, 吴少华, 康建宏, 吴宏亮, 杨生龙, 王星强, 李昱, 黄玉峰. 不同种植模式对水稻籽粒淀粉含量及淀粉关键酶活性的影响[J]. 中国水稻科学, 2024, 38(3): 303-315. |
| [13] | 关雅琪, 鄂志国, 王磊, 申红芳. 影响中国水稻生产环节外包发展因素的实证研究:基于群体效应视角[J]. 中国水稻科学, 2024, 38(3): 324-334. |
| [14] | 许用强, 姜宁, 奉保华, 肖晶晶, 陶龙兴, 符冠富. 水稻开花期高温热害响应机理及其调控技术研究进展[J]. 中国水稻科学, 2024, 38(2): 111-126. |
| [15] | 吕海涛, 李建忠, 鲁艳辉, 徐红星, 郑许松, 吕仲贤. 稻田福寿螺的发生、危害及其防控技术研究进展[J]. 中国水稻科学, 2024, 38(2): 127-139. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||