
Chinese Journal OF Rice Science ›› 2025, Vol. 39 ›› Issue (6): 779-788.DOI: 10.16819/j.1001-7216.2025.250208
• Research Papers • Previous Articles Next Articles
HOU Guihua1, ZHOU Liguo2, LEI Jianguo1, CHEN Hong1, NIE Yuanyuan1,*( )
)
Received:2025-02-24
Revised:2025-03-28
Online:2025-11-10
Published:2025-11-19
Contact:
NIE Yuanyuan
侯桂花1, 周立国2, 雷建国1, 陈虹1, 聂元元1,*( )
)
通讯作者:
聂元元
基金资助:HOU Guihua, ZHOU Liguo, LEI Jianguo, CHEN Hong, NIE Yuanyuan. Preliminary Analysis of Function and Mechanism of OsRDR5 Gene in Rice[J]. Chinese Journal OF Rice Science, 2025, 39(6): 779-788.
侯桂花, 周立国, 雷建国, 陈虹, 聂元元. 水稻OsRDR5基因功能及作用机制初步解析[J]. 中国水稻科学, 2025, 39(6): 779-788.
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.ricesci.cn/EN/10.16819/j.1001-7216.2025.250208
| 用途 Purpose | 引物名称 Primer name | 引物序列(5′-3′) Primer sequence (5′-3′) |
|---|---|---|
| 基因克隆 Gene cloning | OsRDR5-1F | ATGGACGACGACGACCATG |
| OsRDR5-1R | TTAGGATCCCGCGGTGGA | |
| 转基因鉴定 Identification of transgenes | OsRDR5-2F | TATGTTTGTCCGCCCCTCTG |
| OsRDR5-2R | GAGAGAGTGGGGTGGAGGAA | |
| 组织表达模式分析 Analysis of tissue expression patterns | OsRDR5-qRT-3F | CCCTCTATACTGAACCGATGAC |
| OsRDR5-qRT-3R | TAAAACTGCAATCCAACTCTGC | |
| 亚细胞定位 Subcellular localization | OsRDR5-4F | ATGGACGACGACGACCATGACC |
| OsRDR5-4R | AGATCCTCCTCCAGATCCTCCTC | |
| 内参基因Actin1 Reference gene Actin1 | Actin1-F | CATCTATGAAGGATATGCTCTC |
| Actin1-R | CCGTTGTGGTGAATGAGT | |
| LOC_Os01g01430 qRT-PCR | qRT-F | TACTGCTGGGCGTGAAGAAG |
| qRT-R | GCGTCTGCGGCTCAAATATG | |
| LOC_Os03g04070 qRT-PCR | qRT-F | AGTGGTACTTCTTCGTGCCG |
| qRT-R | CGTGACGTCTGCTTCTTCCT | |
| LOC_Os11g03370 qRT-PCR | qRT-F | TATGGAGGCAGCAGCAACAA |
| qRT-R | AGCTACAGGAGGAGGTGGAG | |
| LOC_Os08g37370 qRT-PCR | qRT-F | GAAGTGGACCCAGGAGAACG |
| qRT-R | ATGAACCCCTTGTACAGCGC | |
| LOC_Os11g05614 qRT-PCR | qRT-F | ATCTCCTCCGCCACAAACAG |
| qRT-R | GCCATGATCCGACGATGCTA | |
| LOC_Os01g64310 qRT-PCR | qRT-F | ATTCACCACGAGATGCTGGG |
| qRT-R | GCCACCTTGATCCATCAGCT | |
| LOC_Os02g26720 qRT-PCR | qRT-F | CCTCCAGGAGTTCGTCAACC |
| qRT-R | TTAGACCGATACCCAGCCCA |
Table 1. Primers used in this study
| 用途 Purpose | 引物名称 Primer name | 引物序列(5′-3′) Primer sequence (5′-3′) |
|---|---|---|
| 基因克隆 Gene cloning | OsRDR5-1F | ATGGACGACGACGACCATG |
| OsRDR5-1R | TTAGGATCCCGCGGTGGA | |
| 转基因鉴定 Identification of transgenes | OsRDR5-2F | TATGTTTGTCCGCCCCTCTG |
| OsRDR5-2R | GAGAGAGTGGGGTGGAGGAA | |
| 组织表达模式分析 Analysis of tissue expression patterns | OsRDR5-qRT-3F | CCCTCTATACTGAACCGATGAC |
| OsRDR5-qRT-3R | TAAAACTGCAATCCAACTCTGC | |
| 亚细胞定位 Subcellular localization | OsRDR5-4F | ATGGACGACGACGACCATGACC |
| OsRDR5-4R | AGATCCTCCTCCAGATCCTCCTC | |
| 内参基因Actin1 Reference gene Actin1 | Actin1-F | CATCTATGAAGGATATGCTCTC |
| Actin1-R | CCGTTGTGGTGAATGAGT | |
| LOC_Os01g01430 qRT-PCR | qRT-F | TACTGCTGGGCGTGAAGAAG |
| qRT-R | GCGTCTGCGGCTCAAATATG | |
| LOC_Os03g04070 qRT-PCR | qRT-F | AGTGGTACTTCTTCGTGCCG |
| qRT-R | CGTGACGTCTGCTTCTTCCT | |
| LOC_Os11g03370 qRT-PCR | qRT-F | TATGGAGGCAGCAGCAACAA |
| qRT-R | AGCTACAGGAGGAGGTGGAG | |
| LOC_Os08g37370 qRT-PCR | qRT-F | GAAGTGGACCCAGGAGAACG |
| qRT-R | ATGAACCCCTTGTACAGCGC | |
| LOC_Os11g05614 qRT-PCR | qRT-F | ATCTCCTCCGCCACAAACAG |
| qRT-R | GCCATGATCCGACGATGCTA | |
| LOC_Os01g64310 qRT-PCR | qRT-F | ATTCACCACGAGATGCTGGG |
| qRT-R | GCCACCTTGATCCATCAGCT | |
| LOC_Os02g26720 qRT-PCR | qRT-F | CCTCCAGGAGTTCGTCAACC |
| qRT-R | TTAGACCGATACCCAGCCCA |
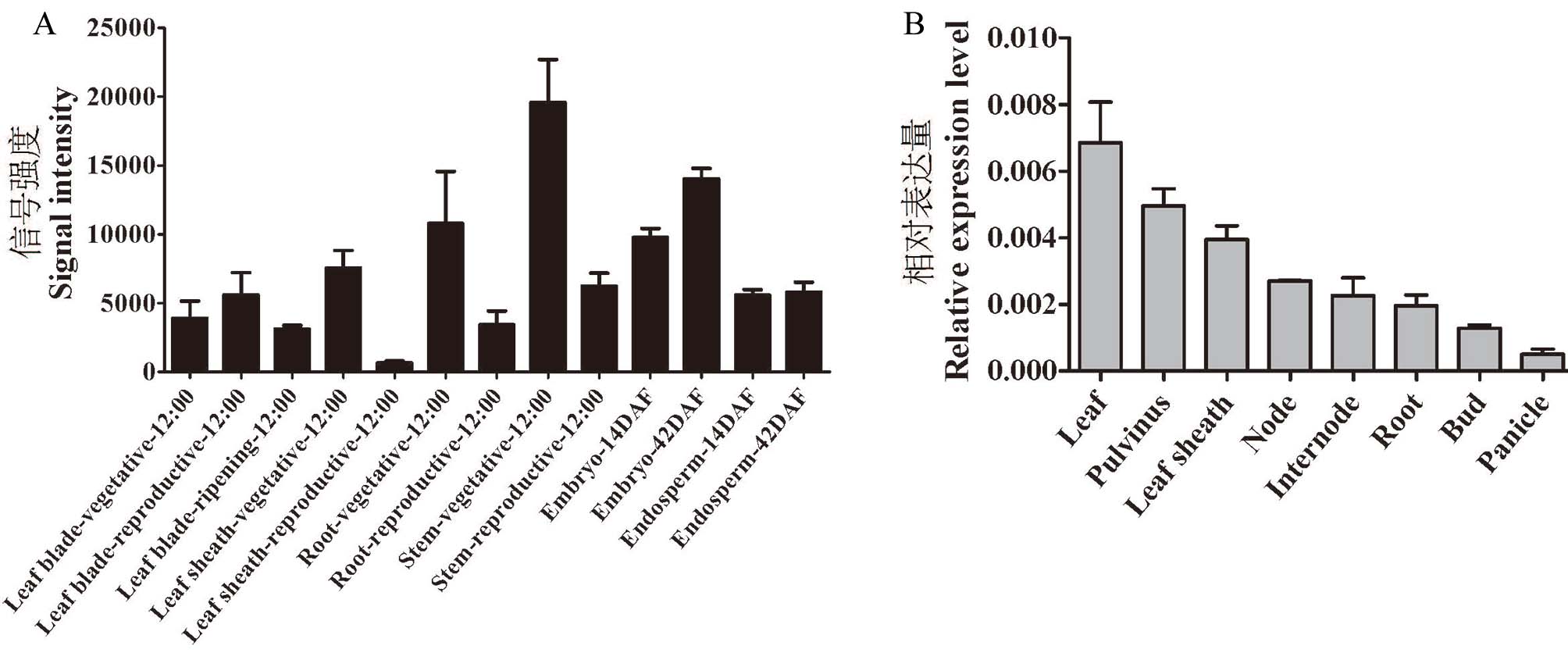
Fig. 5. Tissue-specific expression of OsRDR5 A, OsRDR5 chip data expression level; Signal intensity, OsRDR5 gene signal intensity; Vegetative, Vegetative growth period; Reproductive, Reproductive growth period; Ripening, Ripening period; 14DAF and 42DAF represent embryos and endosperms after 14 days and 42 days of development, respectively; 12:00 indicates tissue sampling time; B, Actin1 gene was used as an internal control. Values are means±SE, n = 3.

Fig. 6. Subcellular localization of OsRDR5 From left to right, the fluorescence channel, marker channel, bright field and overlay images are shown; eGFP, RDR5-eGFP green fluorescent protein; ER, Endoplasmic reticulum; ER marker, SPER-mKATE red fluorescent protein; Bright field, Photographs captured under bright lighting conditions; Merged, An image of the superposition of three channels: eGFP, ER marker and Bright field; Scale bar =10 μm.

Fig. 7. Sequence analysis of OsRDR5 gene A, Analysis of OsRDR5 gene structure and mutation sites. In the OsRDR5 gene sequence, black lines indicate introns, black blocks indicate exons, and black boxes indicate UTRs; WT was the wild type, rdr5-1 and rdr5-2 were two mutants; All the sequences displayed were the target sites and PAM sites; Red horizontal lines in the mutant sequence indicate base deletions; B, Protein sequence alignment analysis. WT encodes 185 amino acids, rdr5-1 encodes 35 amino acids, and rdr5-2 encodes 24 amino acids. Identical amino acids are shaded in blue.

Fig. 8. Ratios of deep rooting of WT and rdr5 mutants A, Deep root ratio and root distribution map of WT and rdr5 lines. The deep root ratio = Number of deep roots/total number of roots; Data are shown as mean ± SD, (n = 12); *,** indicate significance at levels of 0.05, 0.01 by independent t-test, respectively.

Fig. 9. Transcriptome analysis of WT and rdr5 mutant roots A, Differentially expressed genes: P≤0.05 and |log2FC|≥1; Red represents up-regulated genes and blue represents down-regulated genes; B, qRT-PCR verification analysis of differentially expressed genes; The qRT-PCR and RNA-Seq values of the rdr5-1 and rdr5-2 lines were measured using WT as the calibration standard, where the transcriptome sequencing data were calculated using the formula: rdr5-1(FC)=rdr5-1(FPKM)/WT(FPKM), rdr5-2(FC)=rdr5-2(FPKM)/WT(FPKM); C, GO enrichment analysis; D, KEGG enrichment analysis. The abscissa represents the proportion of genes in the corresponding item in all genes of the item, and the ordinate represents different gene function items; The size of the circle represents the number of genes enriched in the corresponding entry, and the larger the circle, the more genes enriched in the pathway; The color represents enrichment significance, the circle indicates that the gene function is associated with both up-regulated and down-regulated genes, the upper triangle indicates that it is only associated with up-regulated genes, the lower triangle indicates that it is only associated with down-regulated genes, and the color represents enrichment significance.
| 基因ID Gene ID | 基因符号 Gene symbol | 表达量差异倍数 log2FC | P值 P-value | 功能 Function |
|---|---|---|---|---|
| LOC_Os01g01430 | OsNAC016 | 1.20 | 0.031 | 负调控水稻抗旱性Negative regulation of rice drought resistance |
| LOC_Os03g04070 | ONAC022 | 1.40 | 0.0002 | 正调控水稻抗旱性Positive regulation of rice drought resistance |
| LOC_Os11g03370 | OsNAC45 | 1.10 | 9.26E-06 | 正调控水稻抗旱性Positive regulation of rice drought resistance |
| LOC_Os01g64310 | ENAC1 | 3.32 | 1.87E-12 | 干旱诱导表达基因Drought-induced expression gene |
Table 2. Differentially expressed genes in response to drought stress
| 基因ID Gene ID | 基因符号 Gene symbol | 表达量差异倍数 log2FC | P值 P-value | 功能 Function |
|---|---|---|---|---|
| LOC_Os01g01430 | OsNAC016 | 1.20 | 0.031 | 负调控水稻抗旱性Negative regulation of rice drought resistance |
| LOC_Os03g04070 | ONAC022 | 1.40 | 0.0002 | 正调控水稻抗旱性Positive regulation of rice drought resistance |
| LOC_Os11g03370 | OsNAC45 | 1.10 | 9.26E-06 | 正调控水稻抗旱性Positive regulation of rice drought resistance |
| LOC_Os01g64310 | ENAC1 | 3.32 | 1.87E-12 | 干旱诱导表达基因Drought-induced expression gene |
| [1] | Luo L J. Breeding for water-saving and drought-resistance rice (WDR) in China[J]. Journal of Experimental Botany, 2010, 61(13): 3509-3517. |
| [2] | Pandey V, Shukla A. Acclimation and tolerance strategies of rice under drought stress[J]. Rice Science, 2015, 22(4): 147-161. |
| [3] | Gupta A, Rico-Medina A, Canõ-Delgado A I. The physiology of plant responses to drought[J]. Science, 2020, 368: 266-269. |
| [4] | 罗利军. 水旱稻分化与节水抗旱稻[J]. 自然杂志, 2022, 44(5): 339-346. |
| Luo L J. Differentiation of lowland-upland rice and development of water-saving and drought-resistant rice[J]. Chinese Journal of Nature, 2022, 44(5): 339-346. (in Chinese with English abstract) | |
| [5] | Farooq M, Wahid A, Lee D J, Ito O, Siddique K H M. Advances in drought resistance of rice[J]. Critical Reviews in Plant Sciences, 2009, 28: 199-217. |
| [6] | Henry A, Cal A J, Batoto T C, Torres R O, Serraj R. Root attributes affecting water uptake of rice (Oryza sativa) under drought[J]. Journal of Experimental Botany, 2012, 63: 4751-4763. |
| [7] | Kim Y, Chung Y S, Lee E, Tripathi P, Heo S, Kim K H. Root response to drought stress in rice (Oryza sativa L.)[J]. International Journal of Molecular Sciences, 2020, 21: 1513. |
| [8] | Chen Y, Shen J, Zhang L, Qi H Y, Yang L J, Wang H Y, Wang J X, Wang Y X, Du H, Tao Z, Zhao T, Deng P C, Shu Q Y, Qian Q, Yu H, Song S Y. Nuclear translocation of OsMFT1 that is impeded by OsFTIP1 promotes drought tolerance in rice[J]. Molecular Plant, 2021, 14: 1297-1311. |
| [9] | Kong X Z, Yu S H, Xiong Y L, Song X Y, Nevescanin-Moreno L, Wei X Q, Rao J L, Zhou H, Bennett M J, Pandey B K, Huang G Q. Root hairs facilitate rice root penetration into compacted layers[J]. Current Biology, 2024, 34: 2039-2048. |
| [10] | Han S C, Wang Y L, Li Y X, Zhu R, Gu Y S, Li J, Guo H F, Ye W, Nabi H G, Yang T, Wang Y M, Liu P L, Duan J Z, Sun X M, Zhang Z Y, Zhang H L, Li Z C, Li J J. The OsNAC41-RoLe1-OsAGAP module promotes root development and drought resistance in upland rice[J]. Molecular Plant, 2024, 17: 1573-1593. |
| [11] | O’Toole J C, Bland W L. Genotypic variation in crop plant root systems[J]. Advances in Agronomy, 1987, 41: 91-145. |
| [12] | Kato Y, Abe J, Kamoshita A, Yamagishi J. Genotypic variation in root growth angle in rice (Oryza sativa L.) and its association with deep root development in upland fields with different water regimes[J]. Plant Soil, 2006, 287: 117-129. |
| [13] | Uga Y, Okuno K, Yano M. Dro1, a major QTL involved in deep rooting of rice under upland field conditions[J]. Journal of Experimental Botany, 2011, 62: 2485-2494. |
| [14] | Uga Y, Sugimoto K, Ogawa S, Rane J, Ishitani M, Hara N, Kitomi Y, Inukai Y, Ono K, Kanno N, Inoue H, Takehisa H, Motoyama R, Nagamura Y, Wu J, Matsumoto T, Takai T, Okuno K, Yano M. Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions[J]. Nature Genetics, 2013, 45: 1097-1102. |
| [15] | Kitomi Y, Kanno N, Kawai S, Mizubayashi T, Fukuoka S, Uga Y. QTLs underlying natural variation of root growth angle among rice cultivars with the same functional allele of DEEPER ROOTING 1[J]. Rice, 2015, 8: 16. |
| [16] | Uddin N, Fukuta Y. A region on chromosome 7 related to differentiation of rice (Oryza sativa L.) between lowland and upland ecotypes[J]. Frontiers in Plant Science, 2020, 11: 1135. |
| [17] | Lou Q J, Chen L, Mei H W, Wei H B, Feng F J, Wang P, Xia H, Li T F, Luo L J. Quantitative trait locus mapping of deep rooting by linkage and association analysis in rice[J]. Journal of Experimental Botany, 2015, 66: 4749-4757. |
| [18] | Lou Q J, Chen L, Mei H W, Xu K, Wei H B, Feng F J, Li T F, Pang X, Shi C, Luo L J, Zhong Y. Root transcriptomic analysis revealing the importance of energy metabolism to the development of deep roots in rice (Oryza sativa L)[J]. Frontiers in Plant Science, 2017, 8: 1314. |
| [19] | Xu K, Lou Q J, Wang D, Li T M, Chen S J, Li T F, Luo L J, Chen L. Overexpression of a novel small auxin-up RNA gene, OsSAUR11, enhances rice deep rootedness[J]. BMC Plant Biology, 2023, 23: 319. |
| [20] | Zhou L G, Liu Z C, Liu Y H, Kong D Y, Li T F, Yu S W, Mei H W, Xu X Y, Liu H Y, Chen L, Luo L J. A novel gene OsAHL1 improves both drought avoidance and drought tolerance in rice[J]. Scientific Reports, 2016, 6: 30264. |
| [21] | 聂元元. 东乡野生稻遗传多样性评价与避旱优异基因资源挖掘[D]. 武汉: 华中农业大学, 2022. |
| Nie Y Y. Evaluation of genetic diversity and mining of excellent drought avoidance genes in Dongxiang wild rice[D]. Wuhan: Huazhong Agricultural University, 2022. (in Chinese with English abstract) | |
| [22] | Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, Xie Y, Shen R, Chen S, Wang Z, Chen Y, Guo J, Chen L, Zhao X, Dong Z, Liu Y G. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants[J]. Molecular Plant, 2015, 8(8): 1274-1284. |
| [23] | Livak K J, Schmittgen T D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method[J]. Methods, 2001, 25(4): 402-408. |
| [24] | Peralta S, Clemente P, Sánchez-Martínez A, Calleja m, Hernández-Sierra R, Matsushima Y, Adán C, Ugalde C, Fernández-Moreno M A, Kaguni L S. Coiled coil domain-containing protein 56 (CCDC56) is a novel mitochondrial protein essential for cytochrome c oxidase function[J]. Journal of Biological Chemistry, 2012, 287(29): 24174-24185. |
| [25] | Wikström M, Krab K, Sharma V. Oxygen activation and energy conservation by cytochrome c oxidase[J]. Chemical Reviews, 2018, 118(5): 2469-2490. |
| [26] | Kadenbach B, Hüttemann M. The subunit composition and function of mammalian cytochrome c oxidase[J]. Mitochondrion, 2015, 24: 64-76. |
| [27] | Kondo M, Pablico P P, Aragones D V, Agbisit R, Abe J, Morita S, Courtois B. Genotypic and environmental variations in root morphology in rice genotypes under upland field conditions[J]. Plant and Soil, 2003, 255: 189-200. |
| [28] | 谢建坤, 胡标林, 万勇, 张弢, 李霞, 刘如龙, 黄运红, 戴亮芳, 罗向东. 东乡普通野生稻与栽培稻苗期抗旱性的比较[J]. 生态学报, 2010, 30(06). |
| Xie J K, Hu B L, Wan Y, Zhang T, Li X, Liu R L, Huang Y H, Dai L F, Luo X D. Comparison of drought resistance between common wild rice and cultivated rice at seedling stage in Dongxiang[J]. Acta Ecologica Sinica, 2010, 30(06). (in Chinese with English abstract) | |
| [29] | Zhou S X, Tian F, Zhu Z F, Fu Y C, Wang X K, Sun C Q. Identification of quantitative trait loci controlling drought tolerance at seedling stage in Chinese Dongxiang common wild rice (Oryza rufipogon Griff.)[J]. Acta Genetica Sinica, 2006, 33(6): 551-8. |
| [30] | Zhang F T, Cui F L, Zhang L X, Wen X F, Luo X D, Zhou Y, Li X, Wan Y, Zhang J, Xie J K. Development and identification of a introgression line with strong drought resistance at seedling stage derived from Oryza sativa L. mating with Oryza rufipogon Griff[J]. Euphytica, 2014, 200(1): 1-7. |
| [31] | 王会民, 唐秀英, 龙起樟, 黄永兰, 芦明, 万建林. 一个东乡野生稻苗期耐旱主效QTL-qDR7的分离鉴定[J]. 分子植物育种, 2021, 19(5): 1569-1577. |
| Wang H M, Tang X Y, Long Q Z, Huang Y L, Lu M, Wan J L. Isolation and identification of a major drought tolerance QTL-qDR7 in Dongxiang wild rice at seedling stage[J]. Molecular Plant Breeding, 2021, 19(5): 1569-1577. (in Chinese with English abstract) | |
| [32] | Uga Y Yamamoto, Kanno N, Kawai S, Mizubayashi T, Fukuoka S. A major QTL controlling deep rooting on rice chromosome 4[J]. Scientific Reports, 2013, 3: 3040. |
| [1] | CHEN Ling, LIN Wenying, LIANG Limei, OUYANG Younan, YE Shenghai, JI Zhijuan. Flowering Habits of Rice and Its Application in Breeding japonica Cytoplasmic Male Sterile Lines [J]. Chinese Journal OF Rice Science, 2025, 39(6): 731-743. |
| [2] | WANG Juan, WU Lijuan, HONG Haibo, YAO Zhiwen, WANG Lei, E Zhiguo. Research Progress on Biological Functions of Ubiquitin-conjugating Enzymes in Rice [J]. Chinese Journal OF Rice Science, 2025, 39(6): 744-750. |
| [3] | TAO Shibo, XU Na, XU Zhengjin, LIU Chang, XU Quan. Cloning of Cold6 Conferring Cold Tolerance in Rice [J]. Chinese Journal OF Rice Science, 2025, 39(6): 751-759. |
| [4] | CHEN Wei, YE Yuanmei, ZHAO Jianhua, FENG Zhiming, CHEN Zongxiang, HU Keming, ZUO Shimin. Modifying Heading Date of Nanjing 46 via CRISPR/Cas9-mediated Genome Editing [J]. Chinese Journal OF Rice Science, 2025, 39(6): 760-770. |
| [5] | WANG Shilin, WU Ting, ZHOU Shiqi, SONG Siming, HU Biaolin. Identification of QTLs for Seed Storability in Dongxiang Wild Rice by Integrating BSA-Seq and QTL Analysis [J]. Chinese Journal OF Rice Science, 2025, 39(6): 789-800. |
| [6] | LU Shuai, TAO Tao, LIU Ran, ZHOU Wenyu, CAO Lei, YANG Qingqing, ZHANG Mingqiu, REN Xinzhe, YANG Zhidi, XU Fuxiang, HUAN Haidong, GONG Yuanhang, ZHANG Haocheng, JIN Sukui, CAI Xiuling, GAO Jiping, LENG Yujia. Identification and Gene Cloning of a Long Sterile Lemma and Small Grain Mutant lsg8 in Rice (Oryza sativa L.) [J]. Chinese Journal OF Rice Science, 2025, 39(6): 813-824. |
| [7] | DENG Huan, LIU Yapei, WANG Chunlian, GUO Wei, CHEN Xifeng, JI Zhiyuan. Mapping Analysis of a New Bacterial Blight Resistance Gene Xa49(t) in Rice [J]. Chinese Journal OF Rice Science, 2025, 39(6): 825-831. |
| [8] | ZHANG Lanlan, LIU Dilin, MA Xiaozhi, HUO Xing, KONG Le, LI Jinhua, FU Chongyun, LIAO Yilong, ZHU Manshan, ZENG Xueqin, LIU Wuge, WANG Feng. Quality Traits Affecting Eating Quality in indica Rice Under Different Nitrogen Application Levels in Early and Late Seasons in South China [J]. Chinese Journal OF Rice Science, 2025, 39(6): 832-846. |
| [9] | BIAN Jinlong, REN Gaolei, QIU Shi, XU Fangfu, HU Zhonglei, ZHANG Hongcheng, WEI Haiyan. Effect of Application Methods of Mixed Controlled-release Nitrogen Fertilizer on Yield and Nitrogen Utilization of Good Taste Quality japonica Rice Under Different Mechanical Transplanting Methods in the Huaibei Region [J]. Chinese Journal OF Rice Science, 2025, 39(6): 847-862. |
| [10] | LI Xing, ZHANG Ruichun, CHEN Ge, XIE Jiaxin, XIAO Zhengwu, CAO Fangbo, CHEN Jiana, HUANG Min. Yield Formation and Photosynthetic Characteristics of Machine-transplanted Late-season Rice with Short Growth Duration [J]. Chinese Journal OF Rice Science, 2025, 39(6): 863-872. |
| [11] | LUO Zizi, ZHANG Dejun, CHEN Dongdong, BI Miao, ZHU Yuhan, HAN Xu, WU Qiang, LI Yuechen. Characteristics of Spatiotemporal Evolution of Thermal Resources in Ratoon Rice at Heading and Grain-filling Stages in Sichuan Basin in 1981−2022 [J]. Chinese Journal OF Rice Science, 2025, 39(6): 873-886. |
| [12] | HAO Wenqian, CAI Xingjing, YANG Haidong, WU Yuyang, TENG Xuan, XUE Chao, GONG Zhiyun. Advances in Roles of Different Types of Histone Modifications in Responses of Rice to Abiotic Stresses [J]. Chinese Journal OF Rice Science, 2025, 39(5): 575-585. |
| [13] | LU Tingting, YAN Wenhui, SU Xinquan, ZENG Luohua, HUA Liqin, CHEN Jianghua, FENG Baohua, WANG Yuexing, HU Jiang, FU Guanfu. Research Progress on Physiological and Ecological Mechanisms and Regulation Pathways of Yield, Quality and Stress Resistance Response in Perennial Rice [J]. Chinese Journal OF Rice Science, 2025, 39(5): 586-600. |
| [14] | WU Wanting, XU Qian, LIU Dantong, ZHU Changjin, DU Haotian, JU Haoran, HUO Zhongyang, DAI Qigen, LI Guohui, XU Ke. Research Progress in Regulation of Anthocyanin Accumulation in Colored Rice [J]. Chinese Journal OF Rice Science, 2025, 39(5): 601-614. |
| [15] | WANG Jingbo, SU Chang, FENG Jing, JIANG Sixu, XU Hai, CUI Zhibo, ZHAO Minghui. Functional Study on Aluminum Tolerance of OsAlR1 Gene in Rice [J]. Chinese Journal OF Rice Science, 2025, 39(5): 615-623. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||