
Chinese Journal OF Rice Science ›› 2020, Vol. 34 ›› Issue (2): 143-149.DOI: 10.16819/j.1001-7216.2020.9085
• Research Papers • Previous Articles Next Articles
Feng MENG, Yaling ZHANG*( ), Xuehui JIN
), Xuehui JIN
Received:2019-07-21
Revised:2019-11-21
Online:2020-03-10
Published:2020-03-10
Contact:
Yaling ZHANG
通讯作者:
张亚玲
基金资助:CLC Number:
Feng MENG, Yaling ZHANG, Xuehui JIN. Detection and Analysis of Magnaporthe oryzae Avirulent Gene AVR-Pita and Its Homologous Genes in Heilongjiang Province[J]. Chinese Journal OF Rice Science, 2020, 34(2): 143-149.
孟峰, 张亚玲, 靳学慧. 黑龙江省稻瘟病菌无毒基因AVR-Pita及其同源基因的检测与分析[J]. 中国水稻科学, 2020, 34(2): 143-149.
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.ricesci.cn/EN/10.16819/j.1001-7216.2020.9085
| 无毒基因 Avr-gene | 前引物序列 Forward primer (5′-3′) | 后引物 Reverse primer (5′-3′) | 目的片段长度 Length of target fragment/bp |
|---|---|---|---|
| AVR-Pita1 | TGCCAATAGACTAGCTTCCG | ATTCCCTCCATTCCAACACT | 1957 |
| AVR-Pita2 | TTTCGGCCCAACTCCGGTCC | TAAAGGGTCCACTGACCCCG | 1642 |
| AVR-Pita3 | AAATATTACCTGCCAGCTGG | CTAAACGAATCGACGCTCCC | 1487 |
Table 1 Primers for Magnaporthe oryzae avirulent gene AVR-Pita amplification.
| 无毒基因 Avr-gene | 前引物序列 Forward primer (5′-3′) | 后引物 Reverse primer (5′-3′) | 目的片段长度 Length of target fragment/bp |
|---|---|---|---|
| AVR-Pita1 | TGCCAATAGACTAGCTTCCG | ATTCCCTCCATTCCAACACT | 1957 |
| AVR-Pita2 | TTTCGGCCCAACTCCGGTCC | TAAAGGGTCCACTGACCCCG | 1642 |
| AVR-Pita3 | AAATATTACCTGCCAGCTGG | CTAAACGAATCGACGCTCCC | 1487 |
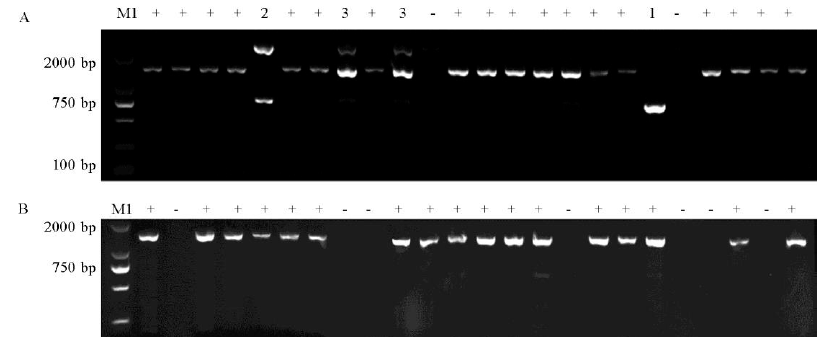
Fig. 1. PCR detection of AVR-Pita and its homologous genes amplification to the tested strains. A, Amplification result of AVR-Pita1; B, Amplification result of AVR-Pita3; M1, Marker DL2000; +, Presence; -, Absence; 1, Low band; 2, Double bands; 3, Three bands.
| 地区Area | 供试菌株数 Number of strains | 无毒基因检出率 Detection rate of avirulence genes /% | ||||
|---|---|---|---|---|---|---|
| AVR-Pita1 | AVR-Pita2 | AVR-Pita3 | ||||
| 齐齐哈尔甘南 Gannan, Qiqihar | 16 | 6.2 | 0 | 81.3 | ||
| 大庆杜蒙Dumeng, Daqing | 19 | 0 | 0 | 73.7 | ||
| 大庆肇源 Zhaoyuan, Dayuan | 11 | 0 | 0 | 100.0 | ||
| 绥化望奎 Wangkui, Suihua | 7 | 42.9 | 0 | 28.6 | ||
| 绥化庆安 Anqing, Suihua | 14 | 42.9 | 0 | 71.4 | ||
| 哈尔滨通河 Tonghe, Harbin | 16 | 31.2 | 0 | 31.2 | ||
| 哈尔滨依兰Yilan, Harbin | 23 | 52.2 | 0 | 52.2 | ||
| 哈尔滨方正Fangzheng, Harbin | 4 | 0 | 0 | 50.0 | ||
| 哈尔滨尚志 Shangzhi, Harbin | 10 | 0 | 0 | 80.0 | ||
| 佳木斯桦川 Huachuan, Jiamusi | 10 | 90.0 | 0 | 80.0 | ||
| 佳木斯汤原 Tangyuan, Jiamusi | 22 | 22.7 | 0 | 50.0 | ||
| 佳木斯桦南 Huanan, Jiamusi | 15 | 86.7 | 0 | 53.3 | ||
| 鹤岗绥滨 Suibin, Hegang | 16 | 68.7 | 0 | 62.5 | ||
| 鸡西虎林 Hulin, Jixi | 9 | 33.3 | 0 | 44.4 | ||
| 鸡西密山 Mishan, Jixi | 10 | 50.0 | 0 | 20.0 | ||
| 合计 | 202 | 36.1 | 0 | 59.4 | ||
Table 2 Distribution of AVR-Pita1 and its homologous genes of Magnaporthe oryzae in Heilongjiang Province in 2017.
| 地区Area | 供试菌株数 Number of strains | 无毒基因检出率 Detection rate of avirulence genes /% | ||||
|---|---|---|---|---|---|---|
| AVR-Pita1 | AVR-Pita2 | AVR-Pita3 | ||||
| 齐齐哈尔甘南 Gannan, Qiqihar | 16 | 6.2 | 0 | 81.3 | ||
| 大庆杜蒙Dumeng, Daqing | 19 | 0 | 0 | 73.7 | ||
| 大庆肇源 Zhaoyuan, Dayuan | 11 | 0 | 0 | 100.0 | ||
| 绥化望奎 Wangkui, Suihua | 7 | 42.9 | 0 | 28.6 | ||
| 绥化庆安 Anqing, Suihua | 14 | 42.9 | 0 | 71.4 | ||
| 哈尔滨通河 Tonghe, Harbin | 16 | 31.2 | 0 | 31.2 | ||
| 哈尔滨依兰Yilan, Harbin | 23 | 52.2 | 0 | 52.2 | ||
| 哈尔滨方正Fangzheng, Harbin | 4 | 0 | 0 | 50.0 | ||
| 哈尔滨尚志 Shangzhi, Harbin | 10 | 0 | 0 | 80.0 | ||
| 佳木斯桦川 Huachuan, Jiamusi | 10 | 90.0 | 0 | 80.0 | ||
| 佳木斯汤原 Tangyuan, Jiamusi | 22 | 22.7 | 0 | 50.0 | ||
| 佳木斯桦南 Huanan, Jiamusi | 15 | 86.7 | 0 | 53.3 | ||
| 鹤岗绥滨 Suibin, Hegang | 16 | 68.7 | 0 | 62.5 | ||
| 鸡西虎林 Hulin, Jixi | 9 | 33.3 | 0 | 44.4 | ||
| 鸡西密山 Mishan, Jixi | 10 | 50.0 | 0 | 20.0 | ||
| 合计 | 202 | 36.1 | 0 | 59.4 | ||
| [1] | Couch B C, Kohn L M.A multilocus gene genealogy concordant with host preference indicates segregation of a new species, Magnaporthe oryzae, from M. grisea[J]. Mycologia, 2002, 94(4): 683-693. |
| [2] | 曹妮, 陈渊, 季芝娟, 曾宇翔, 杨长登, 梁燕. 水稻抗稻瘟病分子机制研究进展[J]. 中国水稻科学, 2019, 33(6): 489-498. |
| Cao N, Chen Y, Ji Z J, Zeng Y X, Yang C D, Liang Y.Recent progress in molecular mechanism of rice blast resistance[J]. Chinese Journal of Rice Science, 2019, 33(6): 489-498. (in Chinese with English abstract) | |
| [3] | Marcel S, Sawers R, Oakeley E, Angliker H, Paszkowski U.Tissue-adapted invasion strategies of the rice blast fungus Magnaporthe oryzae[J]. The Plant Cell, 2010, 22(9): 3177-3187. |
| [4] | Liu W, Zhou X, Li G, Li L, Kong L, Wang C, Zhang H, Xu J.Multiple plant surface signals are sensed by different mechanisms in the rice blast fungus for appressorium formation[J]. PLoS Pathogens, 2011, 7(1): e1001261. |
| [5] | Sweigard J A, Carroll A M, Kang S, Farrall L, Chumley F G, Valent B.Identification, cloning, and character- ization of PWL2, a gene for host species specificity in the rice blast fungus[J]. The Plant Cell, 1995, 7(8): 1221-1233. |
| [6] | Kang S, Sweigard J A, Valent B.The PWL host specificity gene family in the blast fungus Magnaporthe grisea[J]. Molecular Plant-Microbe Interactions, 1995, 8(6): 939-948. |
| [7] | Orbach M J, Farrall L, Sweigard J A, Chumley F G, Valent B.A telomeric avirulence gene determines efficacy for the rice blast resistance gene Pi-ta[J]. The Plant Cell, 2000, 12(11): 2019-2032. |
| [8] | Collemare J, Pianfetti M, Houlle A E, Morin D, Camborde L, Gagey M J, Barbisan C, Fudal I, Lebrun M H, BőHnert H U. Magnaporthe grisea avirulence gene ACE1 belongs to an infection-specific gene cluster involved in secondary metabolism[J]. New Phytologist, 2010, 179(1): 196-208. |
| [9] | Farman M L, Leong S A.Chromosome walking to the AVR1-CO39 avirulence gene of Magnaporthe grisea: Discrepancy between the physical and genetic maps[J]. Genetics, 1998, 150(3): 1049-1058. |
| [10] | Li W, Wang B, Wu J, Lu G, Hu Y, Zhang X, Zhang Z G, Zhao Q, Zhang H Y, Wang Z Y, Wang G L, Han B, Wang Z H, Zhou B.The Magnaporthe oryzae avirulence gene AvrPiz-t encodes a predicted secreted protein that triggers the immunity in rice mediated by the blast resistance gene Piz-t[J]. Molecular Plant-Microbe Interactions, 2009, 22(4): 411-420. |
| [11] | Yoshida K, Saitoh H, Fujisawa S, Kanzaki H, Matsumura H, Yoshida K, Tosa Y, Chuma I, Takano Y, Win J, Kamoun S, Terauchi R.Association genetics reveals three novel avirulence genes from the rice blast fungal pathogen Magnaporthe oryzae[J]. The Plant Cell, 2009, 21(5): 1573-1591. |
| [12] | Wu J, Kou Y, Bao J, Li Y, Tang M, Zhu X, Ponaya A, Xiao G, Li J, Li C, Song M, Cumagun C J, Deng Q, Lu G, Jeon J, Naqvi N I, Zhou B.Comparative genomics identifies the Magnaporthe oryzae avirulence effector Avr-Pi9 that triggers Pi9-mediated blast resistance in rice[J]. New Phytologist, 2015, 206(4): 1463-1475. |
| [13] | Schneider D R, Saraiva A M, Azzoni A R, Miranda H R, Toledo M A, Pelloso A C, Souza A P.Overexpression and purification of PWL2D, a mutant of the effector protein PWL2 from Magnaporthe grisea[J]. Protein Expression and Purification, 2010, 74(1): 24-31. |
| [14] | Zhang S, Wang L, Wu W, He L, Yang X, Pan X.Function and evolution of Magnaporthe oryzae avirulence gene Avr-Pib responding to the rice blast resistance gene Pib[J]. Scientific Reports, 2015, 5: 11642. |
| [15] | Khang C H, Berruyer R, Giraldo M C, Kankanala P, Park S Y, Czymmek K, Kang S, Valent B.Translocation of Magnaporthe oryzae effectors into rice cells and their subsequent cell-to-cell movement[J]. The Plant Cell, 2010, 22(4): 1388-1403. |
| [16] | 甘玉姿, 肖贵, 邓启云, 吴俊, 柏斌, 卢向阳, 周波. 菲律宾稻瘟病菌生理小种中AVR-Pita及其同源基因的序列与功能分析[J]. 中国生物防治学报, 2018, 34(3): 488-498. |
| Gan Y Z, Xiao G, Deng Q Y, Wu J, Bai B, Lu X Y, Zhou B.Sequence and functional analysis of AVR-Pita and its homologous genes in physiological races of Magnaporthe grisea in the Philippines[J]. Chinese Journal of Biological Control, 2018, 34(3): 488-498. (in Chinese with English abstract) | |
| [17] | Chuma I, Isobe C, Hotta Y, Ibaragi K, Futamata N, Kusaba M, Yoshida K, Terauchi R, Fujita Y, Nakayashiki H, Valent B, Tose Y.Multiple translocation of the Avr-pita effector gene among chromosomes of the rice blast fungus Magnaporthe oryzae and related species[J]. PloS Pathogens, 2011, 7(7): 395-396. |
| [18] | 刘殿宇. 黑龙江省稻瘟病菌致病性与无毒基因检测及遗传多样性分析[D]. 大庆: 黑龙江八一农垦大学, 2017. |
| Liu D Y.Pathogenicity avirulent gene detection and genetic diversity analysis of Magnaporthe oryzae in Heilongjiang Province[D]. Daqing: Heilongjiang Bayi Agricultural University, 2017. (in Chinese with English abstract) | |
| [19] | 李思博. 辽宁省稻瘟菌无毒基因及水稻抗瘟基因鉴定[D]. 沈阳:沈阳农业大学, 2018. |
| Li S B.Identification of avirulent genes and resistance genes of Magnaporthe grisea in Liaoning Province[D]. Shenyang: Shenyang Agricultural University, 2018. (in Chinese with English abstract) | |
| [20] | 朱名海, 赵美, 舒灿伟, 周而勋. 南繁区稻瘟病菌无毒基因的检测[J]. 华中农业大学学报, 2017, 36(4): 21-25. |
| Zhu M H, Zhao M, Shu C W, Zhou E X.Detection of avirulence genes in Magnaporthe oryzae from South China Crop Breeding Area[J]. Journal of Huazhong Agricultural University, 2017, 36(4): 21-25. (in Chinese with English abstract) | |
| [21] | 蒋金芬, 韩红萍, 梁友方. 滤纸片法低温冷冻保存菌种的实验室应用[J]. 中国公共卫生, 2006, 22(3): 310. |
| Jiang J F, Han H P, Liang Y F.Laboratory application of filter paper method for cryopreservation[J]. Chinese Public Health, 2006, 22(3): 310-310. (in Chinese) | |
| [22] | 靳学慧. 农业植物病理学[M]. 赤峰: 内蒙古科学技术出版社,1999. |
| Jin X H.Agricultural Plant Pathology[M]. Chifeng: Inner Mongolia Science and Technology Press, 1999. (in Chinese) | |
| [23] | Zhou E, Jia Y, Singh P, Correll J C, Lee F N.Instability of the Magnaporthe oryzae avirulence gene AVR-Pita alters virulence[J]. Fungal Genetics and Biology, 2007, 44(10): 1034. |
| [24] | 余欢, 姜华, 王艳丽, 孙国昌. 无毒基因在不同寄主梨孢菌中的变异研究[J]. 浙江农业学报, 2015, 27(8): 1414-1421. |
| Yu H, Jiang H, Wang Y L, Sun G C.Study on the variation of avirulence genes in different host Phasporium[J]. Acta Agriculturae Zhejiangensis, 2015, 27(8): 1414-1421. (in Chinese with English abstract) | |
| [25] | Dai Y, Jia Y, Correll J, Wang X, Wang Y.Diversification and evolution of the avirulence gene AVR-Pita1 in field isolates of Magnaporthe oryzae[J]. Fungal Genetics and Biology, 2010, 47(12): 973-980. |
| [26] | Chuma I, Isobe C, Hotta Y, Ibaragi K, Kusaba M, Yoshida K, Terauchi R, Fujita Y, Nakayashiki H, Valent B, Tosa Y.Multiple Translocation of the AVR-Pita effector gene among chromosomes of the rice blast fungus Magnaporthe oryzae and related species[J]. PloS Pathogens, 2011, 7(7): 395-396. |
| [27] | 于连鹏. 黑龙江省主栽水稻品种Pita、Pia和Piz-t抗瘟基因检测和抗性评价[D]. 大庆: 黑龙江八一农垦大学, 2017. |
| Yu L P.Pita, Pia and Piz-t genes detection and blast resistance evaluation of main rice varieties in Heilongjiang Province[D]. Daqing: Heilongjiang Bayi Agricultural University, 2017. (in Chinese with English abstract) | |
| [28] | 周江鸿, 王久林, 蒋琬如, 雷财林, 凌忠专.我国稻瘟病菌毒力基因的组成及其地理分布[J]. 作物学报, 2003(5): 646-651. |
| Zhou J H, Wang J L, Jiang W R, Lei C L, Ling Z Z.Composition and geographical distribution of virulence genes of rice blast fungus in China[J]. Acta Agronomica Sinica, 2003(5): 646-651. (in Chinese with English abstract) |
| [1] | Shufang LIU, Liying DONG, Xundong LI, Wumin ZHOU, Qinzhong YANG. Different Reactions of Rice Monogenic Line IRBL9-W Harboring Pi9 Gene to Magnaporthe oryzae Containing AvrPi9 During Seedling and Adult-plant Stages [J]. Chinese Journal OF Rice Science, 2021, 35(3): 303-310. |
| [2] | Xiangyang FENG, Zhen ZHANG, Rongyao CHAI, Haiping QIU, Jiaoyu WANG, Xueqin MAO, Yanli WANG, Guochang SUN. Functional Analysis of MoMET3 in Growth, Development and Pathogenicity of Magnaporthe oryzae [J]. Chinese Journal OF Rice Science, 2017, 31(5): 542-550. |
| [3] | Minghai ZHU, Lei PI, Canwei SHU, Erxun ZHOU. AFLP Analyses of Genetic Diversity and Population Genetic Structure of Magnaporthe oryzae from South China Crop Breeding Area [J]. Chinese Journal OF Rice Science, 2017, 31(3): 320-326. |
| [4] | Hongchun RUAN, Niuniu SHI, Yixin DU, Lin GAN, Xiujuan YANG, Yuli DAI, Furu CHEN. Analysis on Resistance of Pi Genes to Predominant Races of Mangnaporthe oryzae in Fujian Province, China [J]. Chinese Journal OF Rice Science, 2017, 31(1): 105-110. |
| [5] | Zhuo-kan GU, Ling LI, Jiao-yu WANG, Rong-yao CHAI, Yan-li WANG, Zhen ZHANG, Xue-qin MAO, Hai-ping QIU, Guo-chang SUN. Observation of Sexual Structure of Magnaporthe oryzae via Calcofluor White and Nile Red Staining [J]. Chinese Journal OF Rice Science, 2016, 30(6): 668-672. |
| [6] | Xin LIU, Heng ZHANG, Hu-fei KAN, Li-shuai ZHOU, Hao HUANG, Lin-lin SONG, Huan-chen ZHAI, Jun ZHANG, Guo-dong LU. Bioinformatic and Expression Analysis of Rice Ubiquitin-conjugating Enzyme Gene Family [J]. Chinese Journal OF Rice Science, 2016, 30(3): 223-231. |
| [7] | Ling WANG, Shi-min ZUO, Ya-fang ZHANG, Zong-xiang CHEN, Xue-biao PAN, Shi-wen HUANG. Genetic Structure of Rice Blast Pathogen Magnaporthe oryzae in Sichuan Province [J]. Chinese Journal OF Rice Science, 2015, 29(3): 327-334. |
| [8] | He LI, Yi-juan HAN, Yi-juan LIN, Li-hua LIU, Cheng-kang ZHANG, Lian-hu ZHANG, Zong-hua WANG, Guo-dong LU. Functional Analysis of Zinc Finger Protein Gene OsZFP1 in Rice [J]. Chinese Journal OF Rice Science, 2015, 29(2): 135-140. |
| [9] | Ya-ping CHEN, Wen-xiao SHI, Hong-kai WANG. Construction of a Vector for Large DNA Fragment Transformation in Magnaporthe oryzae [J]. Chinese Journal OF Rice Science, 2015, 29(1): 91-96. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||