
Chinese Journal OF Rice Science ›› 2022, Vol. 36 ›› Issue (4): 348-356.DOI: 10.16819/j.1001-7216.2022.210604
• Research Papers • Previous Articles Next Articles
ZHOU Yonglin1,2, SHEN Xiaolei1,2, ZHOU Lishuai1,2, LIN Qiaoxia1,2, WANG Zhaolu1,2, CHEN Jing1,2, FENG Huijie1, ZHANG Zhenwen1, CHEN Xiaoting1,2,3,*( ), LU Guodong2
), LU Guodong2
Received:2021-06-10
Revised:2021-09-30
Online:2022-07-10
Published:2022-07-12
Contact:
CHEN Xiaoting
周永林1,2, 申小磊1,2, 周立帅1,2, 林巧霞1,2, 王朝露1,2, 陈静1,2, 冯慧捷1, 张振文1, 陈晓婷1,2,3,*( ), 鲁国东2
), 鲁国东2
通讯作者:
陈晓婷
基金资助:ZHOU Yonglin, SHEN Xiaolei, ZHOU Lishuai, LIN Qiaoxia, WANG Zhaolu, CHEN Jing, FENG Huijie, ZHANG Zhenwen, CHEN Xiaoting, LU Guodong. OsLOX10 Positively Regulates Defense Responses of Rice to Rice Blast and Bacterial Blight[J]. Chinese Journal OF Rice Science, 2022, 36(4): 348-356.
周永林, 申小磊, 周立帅, 林巧霞, 王朝露, 陈静, 冯慧捷, 张振文, 陈晓婷, 鲁国东. OsLOX10正调控水稻对稻瘟病和白叶枯病的抗性[J]. 中国水稻科学, 2022, 36(4): 348-356.
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.ricesci.cn/EN/10.16819/j.1001-7216.2022.210604
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 用途 Purpose | 片段长度Length/bp |
|---|---|---|---|
| OsLOX10-F | ATGCAGCAGCCGCAGGCGAG | 过表达载体构建 Construction of OE vector | 2607 |
| OsLOX10-R | TCAGATGGAAGCGCTGTTGG | ||
| UbiP-seq | TTTTAGCCCTGCCTTCATACGC | 过表达载体测序 Sequencing of OE vector | 2795 |
| NosR-seq | AGACCGGCAACAGGATTCAATC | ||
| OsLOX10-U3-F | ggcaGCTACACCCAACTCCCTACG | CRISPR/Cas9第1靶点验证 Verification of first-target site of CRISPR/Cas9 | 259 |
| OsLOX10-U3-R | aaacCGTAGGGAGTTGGGTGTAGC | ||
| OsLOX10-U6a-F | gccgCGCCAGAGTCTGATCAACGC | CRISPR/Cas9第2靶点验证 Verification of second-target site of CRISPR/Cas9 | 329 |
| OsLOX10-U6a-R | aaacGCGTTGATCAGACTCTGGCG | ||
| U-F | CTCCGTTTTACCTGTGGAATCG | 敲除靶点第1轮扩增 First round amplification of knockout targets | 534 |
| gRNA-R | CGGAGGAAAATTCCATCCAC | ||
| Uctcg-B1' | TTCAGAggtctcTctcgCACTGGAATCGGCAGCAAAGG | CRISPR/Cas9第1靶点扩增 Amplification of first-target site | 564 |
| gRctga-B2 | AGCGTGggtctcGtcagGGTCCATCCACTCCAAGCTC | ||
| Uctga-B2' | TTCAGAggtctcTctgaCACTGGAATCGGCAGCAAAGG | CRISPR/Cas9第2靶点扩增 Amplification of second-target site target site | 629 |
| gRcggt-BL | AGCGTGggtctcGaccgGGTCCATCCACTCCAAGCTC | ||
| Actin-QF | GAGTATGATGAGTCGGGTCCAG | Actin定量PCR qRT-PCR of Actin | 143 |
| Actin-QR | ACACCAACAATCCCAAACAGAG | ||
| OsPR1a-QF | CGTCTTCATCACCTGCAACTACTC | OsPR1a定量PCR qRT-PCR of OsPR1a | 132 |
| OsPR1a-QR | CATGCATAAACACGTAGCATAGCA | ||
| OsPR1b-QF | ACGGGCGTACGTACTGGCTA | OsPR1b定量PCR qRT-PCR of OsPR1b | 105 |
| OsPR1b-QR | CTCGGTATGGACCGTGAAG | ||
| OsAOS2-QF | CAATACGTGTACTGGTCGAATGG | OsAOS2定量PCR qRT-PCR of OsAOS2 | 134 |
| OsAOS2-QR | AAGGTGTCGTACCGGAGGAA | ||
| OsPAL1-QF | AGGAGCTCGGCTGCGTATT | OsPAL1定量PCR qRT-PCR of OsPAL1 | 79 |
| OsPAL1-QR | ATGCCGAGGAACACCTTGTT | ||
| OsLOX5-QF | CTGATGAGGAGTTTGCACGA | OsLOX5定量PCR qRT-PCR of OsLOX5 | 542 |
| OsLOX5-QR | TCGTCCTTCAGGAGCAGAAT | ||
| OsAOC-QF | CCAAGGTGCAGGAGATGTT | OsAOC定量PCR qRT-PCR of OsAOC | 150 |
| OsAOC-QR | TACAGCTTGTTGGTGAAGGG | ||
| OsJAZ-QF | GAAGGCTCAACAGCTGACCAT | OsJAZ定量PCR qRT-PCR of OsJAZ | 69 |
| OsJAZ-QR | TTGGTGGACGGGAAGTTCTC | ||
| OsUG-F | TTCTGGTCCTTCCACTTTCAG | 泛素融合蛋白定量PCR qRT-PCR of ubiquitin fusion protein OsUG | 92 |
| OsUG-R | ACGATTGATTTAACCAGTCCATGA | ||
| MoPot2-F | ACGACCCGTCTTTACTTATTTGG | MoPot2定量PCR qRT-PCR of MoPot2 | 99 |
| MoPot2-R | AAGTAGCGTTGGTTTTGTTGGAT |
Table 1. Primers used in this study.
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 用途 Purpose | 片段长度Length/bp |
|---|---|---|---|
| OsLOX10-F | ATGCAGCAGCCGCAGGCGAG | 过表达载体构建 Construction of OE vector | 2607 |
| OsLOX10-R | TCAGATGGAAGCGCTGTTGG | ||
| UbiP-seq | TTTTAGCCCTGCCTTCATACGC | 过表达载体测序 Sequencing of OE vector | 2795 |
| NosR-seq | AGACCGGCAACAGGATTCAATC | ||
| OsLOX10-U3-F | ggcaGCTACACCCAACTCCCTACG | CRISPR/Cas9第1靶点验证 Verification of first-target site of CRISPR/Cas9 | 259 |
| OsLOX10-U3-R | aaacCGTAGGGAGTTGGGTGTAGC | ||
| OsLOX10-U6a-F | gccgCGCCAGAGTCTGATCAACGC | CRISPR/Cas9第2靶点验证 Verification of second-target site of CRISPR/Cas9 | 329 |
| OsLOX10-U6a-R | aaacGCGTTGATCAGACTCTGGCG | ||
| U-F | CTCCGTTTTACCTGTGGAATCG | 敲除靶点第1轮扩增 First round amplification of knockout targets | 534 |
| gRNA-R | CGGAGGAAAATTCCATCCAC | ||
| Uctcg-B1' | TTCAGAggtctcTctcgCACTGGAATCGGCAGCAAAGG | CRISPR/Cas9第1靶点扩增 Amplification of first-target site | 564 |
| gRctga-B2 | AGCGTGggtctcGtcagGGTCCATCCACTCCAAGCTC | ||
| Uctga-B2' | TTCAGAggtctcTctgaCACTGGAATCGGCAGCAAAGG | CRISPR/Cas9第2靶点扩增 Amplification of second-target site target site | 629 |
| gRcggt-BL | AGCGTGggtctcGaccgGGTCCATCCACTCCAAGCTC | ||
| Actin-QF | GAGTATGATGAGTCGGGTCCAG | Actin定量PCR qRT-PCR of Actin | 143 |
| Actin-QR | ACACCAACAATCCCAAACAGAG | ||
| OsPR1a-QF | CGTCTTCATCACCTGCAACTACTC | OsPR1a定量PCR qRT-PCR of OsPR1a | 132 |
| OsPR1a-QR | CATGCATAAACACGTAGCATAGCA | ||
| OsPR1b-QF | ACGGGCGTACGTACTGGCTA | OsPR1b定量PCR qRT-PCR of OsPR1b | 105 |
| OsPR1b-QR | CTCGGTATGGACCGTGAAG | ||
| OsAOS2-QF | CAATACGTGTACTGGTCGAATGG | OsAOS2定量PCR qRT-PCR of OsAOS2 | 134 |
| OsAOS2-QR | AAGGTGTCGTACCGGAGGAA | ||
| OsPAL1-QF | AGGAGCTCGGCTGCGTATT | OsPAL1定量PCR qRT-PCR of OsPAL1 | 79 |
| OsPAL1-QR | ATGCCGAGGAACACCTTGTT | ||
| OsLOX5-QF | CTGATGAGGAGTTTGCACGA | OsLOX5定量PCR qRT-PCR of OsLOX5 | 542 |
| OsLOX5-QR | TCGTCCTTCAGGAGCAGAAT | ||
| OsAOC-QF | CCAAGGTGCAGGAGATGTT | OsAOC定量PCR qRT-PCR of OsAOC | 150 |
| OsAOC-QR | TACAGCTTGTTGGTGAAGGG | ||
| OsJAZ-QF | GAAGGCTCAACAGCTGACCAT | OsJAZ定量PCR qRT-PCR of OsJAZ | 69 |
| OsJAZ-QR | TTGGTGGACGGGAAGTTCTC | ||
| OsUG-F | TTCTGGTCCTTCCACTTTCAG | 泛素融合蛋白定量PCR qRT-PCR of ubiquitin fusion protein OsUG | 92 |
| OsUG-R | ACGATTGATTTAACCAGTCCATGA | ||
| MoPot2-F | ACGACCCGTCTTTACTTATTTGG | MoPot2定量PCR qRT-PCR of MoPot2 | 99 |
| MoPot2-R | AAGTAGCGTTGGTTTTGTTGGAT |
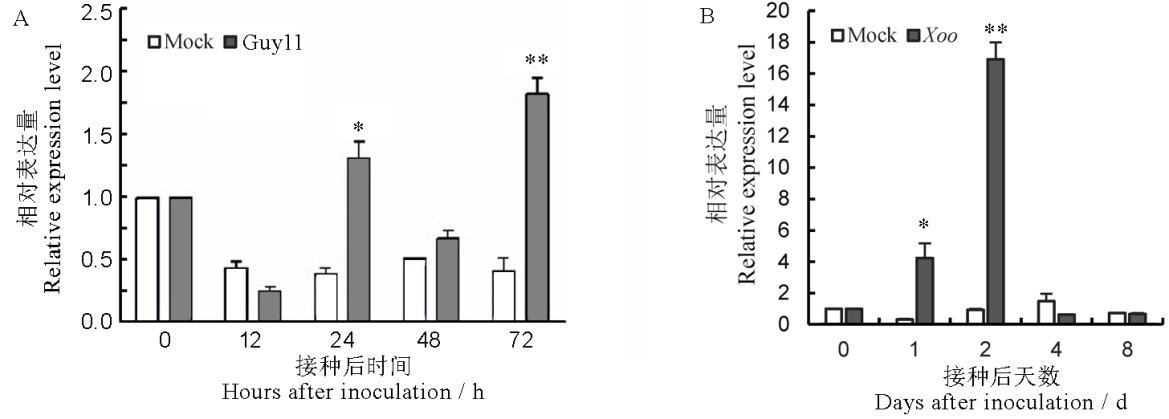
Fig. 1. Expression profiles of OsLOX10 after inoculation with M. oryzae (A) and Xoo (B) in rice. Data in the figure are Mean±SE, and *, ** represent significant difference at the 0.05 and 0.01 levels (t-test), respectively.

Fig. 2. Identification of homozygous mutant of OsLOX10 by gene editing. A, Schematic presentation of the OsLOX10 structure and gene editing site; B, Mutation site in OsLOX10 gene editing line-18; C, Mutation sites in OsLOX10 gene editing line-24.

Fig. 8. Response of OsLOX10-KO transgenic rice and wild-type Nipponbare to flg22 and chitin. A, Chitin-induced ROS burst in the transgenic rice OsLOX10-KO and Nipponbare (rice leaf disks were exposed to 500 nmol/L chitin and water); B, flg22-induced ROS burst in the transgenic rice OsLOX10-KO and NPB (rice leaf disks were exposed to 400 nmol/L flg22 and water).
| [1] | Khush G S. What it will take to feed 5.0 billion rice consumers in 2030[J]. Plant Molecular Biology, 2005, 59(1): 1-6. |
| [2] | Ke Y, Deng H, Wang S. Advances in understanding broad-spectrum resistance to pathogens in rice[J]. The Plant Journal, 2017, 90: 738-748. |
| [3] | Liu W, Wang G L. Plant innate immunity in rice: A defense against pathogen infection[J]. National Science Review, 2016, 3: 295-308 |
| [4] | Ngou B P M, Ahn H K, Ding P, Jones J D G. Mutual potentiation of plant immunity by cell-surface and intracellular receptors[J]. Nature, 2021: 1-6. |
| [5] | Yuan M, Jiang Z, Bi G, Nomura K, Liu M, Wang Y, Cai B, Zhou J M, He S Y, Xin X F. Pattern-recognition receptors are required for NLR-mediated plant immunity[J]. Nature, 2021: 1-5. |
| [6] | Umate P. Genome-wide analysis of lipoxygenase gene family in Arabidopsis and rice[J]. Plant Signaling & Behavior, 2011, 6(3): 335-338. |
| [7] | Marla S S, Singh V K. LOX genes in blast fungus (Magnaporthe grisea) resistance in rice[J]. Functional & Integrative Genomics, 2012, 12(2): 265-275. |
| [8] | Mizuno K, Iida T, Takano A, Yokoyama M, Fujimura T. A new 9-lipoxygenase cDNA from developing rice seeds[J]. Plant Cell Physiology, 2003, 44: 1168-1175. |
| [9] | Shirasawa K, Takeuchi Y, Ebitani T, Suzuki Y. Identification of gene for rice (Oryza sativa) seed lipoxygenase-3 involved in the generation of stale flavor and development of SNP markers for lipoxygenase-3 deficiency[J]. Breeding Science, 2008, 58: 169-176. |
| [10] | Gayen D, Ali N, Ganguly M, Paul S, Datta S K. RNAi mediated silencing of lipoxygenase gene to maintain rice grain quality and viability during storage[J]. Plant Cell Tissue and Organ Culture, 2014, 118(2): 229-243. |
| [11] | Gayen D, Ali N, Sarkar S N, Datta S K, Datta K. Down-regulation of lipoxygenase gene reduces degradation of carotenoids of golden rice during storage[J]. Planta, 2014, 242(1): 353-363. |
| [12] | Ma L, Zhu F, Li Z, Zhang J, Li X, Dong J, Wang T. TALEN-based mutagenesis of lipoxygenase LOX3 enhances the storage tolerance of rice (Oryza sativa) seeds[J]. PLoS ONE, 2015, 10(12): e0143877. |
| [13] | Xu H, Wei Y, Zhu Y, Lian L, Xie H, Cai Q, Chen Q, Lin Z, Xie H, Zhang J. Antisense suppression of LOX3 gene expression in rice endosperm enhances seed longevity[J]. Plant Biotechnology Journal, 2015, 13: 526-539. |
| [14] | Huang J, Cai M, Long Q, Liu L, Lin Q, Jiang L, Chen S, Wan J. OsLOX2, a rice type I lipoxygenase, confers opposite effects on seed germination and longevity[J]. Transgenic Research, 2014, 23(4): 643-655 |
| [15] | Ohta H, Shida K, Peng Y L, Furusawa I, Aibara S, Morita Y. A lipoxygenase pathway is activated in rice after infection with the rice blast fungus Magnaporthe grisea[J]. Plant Physiology, 1991, 97: 94-98. |
| [16] | Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, Xie Y, Shen R, Chen S, Wang Z, Chen Y, Guo J, Chen L, Zhao X, Dong Z, Liu Y G. A robust CRISPR/Cas9 system for convenient, high- efficiency multiplex genome editing in monocot and dicot plants[J]. Molecular Plant, 2015, 8(8): 1274-1284. |
| [17] | Chen S, Songkumarn P, Liu J, Wang G L. A versatile zero background T-vector system for gene cloning and functional genomics[J]. Plant Physiology, 2009, 150(3): 1111-1121. |
| [18] | Hong Y, Liu Q, Cao Y, Zhang Y, Cheng S, Cao L. The OsMPK15 negatively regulates Magnaporthe oryza and Xoo disease resistance via SA and JA signaling pathway in rice[J]. Frontiers in Plant Science, 2019, 10: 752 |
| [19] | Wang G, Ding X, Yuan M, Qiu D, Li X, Xu C, Wang S. Dual function of rice OsDR8 gene in disease resistance and thiamine accumulation[J]. Plant Molecular Biology, 2006, 60: 437-449. |
| [20] | Qiu D, Xiao J, Ding X, Xiong M, Cai M, Li X, Xu C, Wang S. OsWRKY13 mediates rice disease resistance by regulating defense-related genes in salicylate- and jasmonate-dependent signaling[J]. Molecular Plant- Microbe Interactions, 2007, 20(5): 492-499. |
| [21] | Qiu D, Xiao J, Xie W, Liu H, Li X, Wang S. rice gene network inferred from expression profiling of plants overexpressing oswrky13, a positive regulator of disease resistance[J]. Molecular Plant, 2008, 1(3): 538-551. |
| [22] | Tao Z, Liu H, Qiu D, Zhou Y, Xu C, Wang S. A pair of allelic WRKY genes play opposite roles in rice-bacteria interactions[J]. Plant Physiology, 2009, 151: 936-948. |
| [23] | Harkenrider M, Sharma R, de Vleesschauwer D, Tsao L, Zhang X, Chern M, Canlas P, Zuo S, Ronald P C. Overexpression of rice wall-associated kinase25 (OsWAK25) alters resistance to bacterial and fungal pathogens[J]. PLoS ONE, 2016, 11: e0147310. |
| [24] | Tonnessen B W, Manosalva P, Lang J M, Baraoidan M, Bordeos A, Mauleon R, Oard J, Leung H, Leach J E. Rice phenylalanine ammonialyase gene OsPAL4 is associated with broad spectrum disease resistance[J]. Plant Molecular Biology, 2015, 87: 273-286. |
| [25] | Dubouzet J G, Maeda S, Sugano S, Ohtake M, Hayashi N, Ichikawa T, Kondou Y, Horii Y, Matsui M, Hirochika H, Takatsuji H, Mori M. Screening for resistance against Pseudomonas syringae in rice-foxarabidopsis lines identified a putative receptor-like cytoplasmic kinase gene that confers resistance to major bacterial and fungal pathogens in Arabidopsis and rice[J]. Plant Biotechnology Journal, 2010, 9: 466-485. |
| [26] | Zhao X, Zhang T, Feng H, Qiu T, Zhao W. OsNBL1, a multi-organelle localized protein, plays essential roles in rice senescence, disease resistance, and salt tolerance[J]. Rice, 2021, 14(1): 10. |
| [27] | Narayanan S P, Lung S C, Liao P, Lo C, Chye M L. The Overexpression of Osacbp5 protects transgenic rice against necrotrophic, hemibiotrophic and biotrophic pathogens[J]. Scientific Reports, 2020, 10(1): 14918. |
| [28] | Han B, Zhou Y, Zhou Z H, Sun B, Zhou F, Yin C X, Ma W H, Chen H, Lin Y J. Repressed OSMESL expression-triggers reactive oxygen species mediated broad-spectrum disease resistance in rice[J]. Plant Biotechnology Journal, 2021, 19(8): 1511-1522 |
| [29] | Mei C, Qi M, Sheng G, Yang Y Y. inducible overexpression of a rice allele oxide synthase gene increases the endogenous jasmonic acid level, PR gene expression, and host resistance to fungal infection[J]. Molecular Plant Microbe Interactions, 2006, 19(10): 1127-1137. |
| [30] | Nalam V J, Alam S, Keereetaweep J, Shah J. Facilitation of Fusarium graminearum infection by 9-lipoxygenases in Arabidopsis and wheat[J]. Molecular Plant Microbe Interactions, 2015, 28(10): 1142-1152 |
| [31] | Christensen S A, Nemchenko A, Borrego E, Murray I, Sobhy I S, Bosak L, Deblasio S, Vaughn K A, Herrfurth C, Tumlinson J, Nansen C, Meeley R, Kolomiets M V. The maize lipoxygenase, zmlox10, mediates green leaf volatile, jasmonate and-herbivore-induced plant volatile production for defense against insect attack[J]. Plant Journal, 2013, 74(1): 59-73. |
| [32] | Xing Q J, Liao J J, Cao S X, Li M, Lü T H, Qi H Y. Cmlox10 positively regulates drought tolerance through jasmonic acid-mediated stomatal closure in oriental melon (Cucumis melo var. Makuwa Makino)[J]. Scientific Reports, 2020, 10: 17452 |
| [33] | Torres M A, Jones J D, Dangl J L. Reactive oxygen species signaling in response to pathogens[J]. Plant Physiology, 2006, 141: 373-378. |
| [1] | XU Danjie, LIN Qiaoxia, LI Zhengkang, ZHUANG Xiaoqian, LING Yu, LAI Meiling, CHEN Xiaoting, LU Guodong. OsOPR10 Positively Regulates Rice Blast and Bacterial Blight Resistance [J]. Chinese Journal OF Rice Science, 2024, 38(4): 364-374. |
| [2] | CHEN Mingliang, XIONG Wentao, SHEN Yumin, XIONG Huanjin, LUO Shiyou, WU Xiaoyan, HU Lanxiang, XIAO Yeqing. Genetic Dissection of Broad Spectrum Resistance of the Rice Maintainer Ganxiang B [J]. Chinese Journal OF Rice Science, 2023, 37(5): 470-477. |
| [3] | LI Jingfang, WEN Shuyue, ZHAO Lijun, CHEN Tingmu, ZHOU Zhenling, SUN Zhiguang, LIU Yan, CHEN Haiyuan, ZHANG Yunhui, CHI Ming, XING Yungao, XU Bo, XU Dayong, WANG Baoxiang. Development of Aromatic Salt-tolerant Rice Based on CRISPR/Cas9 Technology [J]. Chinese Journal OF Rice Science, 2023, 37(5): 478-485. |
| [4] | LI Gang, GAO Qingsong, LI Wei, ZHANG Wenxia, WANG Jian, CHEN Baoshan, WANG Di, GAO Hao, XU Weijun, CHEN Hongqi, JI Jianhui. Directed Knockout of SD1 Gene Improves Lodging Resistance and Blast Resistance of Rice [J]. Chinese Journal OF Rice Science, 2023, 37(4): 359-367. |
| [5] | DUAN Min, XIE Liujie, GAO Xiuying, TANG Haijuan, HUANG Shanjun, PAN Xiaobiao. Creation of Thermo-sensitive Genic Male Sterile Rice Lines with Wide Compatibility Based on CRISPR/Cas9 Technology [J]. Chinese Journal OF Rice Science, 2023, 37(3): 233-243. |
| [6] | WANG Shiguang, LU Zhanhua, LIU Wei, LU Dongbai, WANG Xiaofei, FANG Zhiqiang, WU Haoxiang, HE Xiuying. Generating Guangdong Simiao Rice Germplasms by Applying CRISPR/Cas9 Gene Editing and Marker-assisted Selection Technology [J]. Chinese Journal OF Rice Science, 2023, 37(1): 29-36. |
| [7] | ZHANG Yuanye, YIN Liying, LI Rongtian, HE Mingliang, LIU Xinxin, PAN Tingting, TIAN Xiaojie, BU Qingyun, LI Xiufeng. Breeding of Rc Function Restoration Red Rice via CRISPR/Cas9 Mediated Genome Editing [J]. Chinese Journal OF Rice Science, 2022, 36(6): 572-578. |
| [8] | YIN Liying, ZHANG Yuanye, LI Rongtian, HE Mingliang, WANG Fangquan, XU Yang, LIU Xinxin, PAN Tingting, TIAN Xiaojie, BU Qingyun, LI Xiufeng. Improvement of Herbicide Resistance in Rice by Using CRISPR/Cas9 System [J]. Chinese Journal OF Rice Science, 2022, 36(5): 459-466. |
| [9] | LI Zhaowei, SUN Congying, LING Donglan, ZENG Huiling, ZHANG Xiaomei, FAN Kai, LIN Wenxiong. Construction of osarf7 Mutants in Rice Based on CRISPR/Cas9 Technology and Investigation on Their Agronomic Traits [J]. Chinese Journal OF Rice Science, 2022, 36(3): 237-247. |
| [10] | LIANG Minmin, ZHANG Huali, CHEN Junyu, DAI Dongqing, DU Chengxing, WANG Huimei, MA Liangyong. Developing Fragrant Early indica TGMS Line with Blast Resistance by Using CRISPR/Cas9 Technology [J]. Chinese Journal OF Rice Science, 2022, 36(3): 248-258. |
| [11] | Yudong CAO, Xiangyi XIAO, Naizhong YE, Xiaowen DING, Xiaoxuan YI, Jinling LIU, Yinghui XIAO. Auxin Regulator OsGRF4 Simultaneously Regulates Rice Grain Shape and Blast Resistance [J]. Chinese Journal OF Rice Science, 2021, 35(6): 629-638. |
| [12] | Kai LU, Tao CHEN, Shu YAO, Wenhua LIANG, Xiaodong WEI, Yadong ZHANG, Cailin WANG. Functional Analysis on Four Receptor-like Protein Kinases Under Salt Stress in Rice [J]. Chinese Journal OF Rice Science, 2021, 35(2): 103-111. |
| [13] | Tianshun ZHOU, Dong YU, Ling LIU, Ning OUYANG, Guilong YUAN, Meijuan DUAN, Dingyang YUAN. CRISPR/Cas9-mediatedEditing of AFP1Improves Rice Stress Tolerance [J]. Chinese Journal OF Rice Science, 2021, 35(1): 11-18. |
| [14] | Mengzhu LI, Gaopeng WANG, Yue WU, Yi REN, Ganghua LI, Zhenghui LIU, Yanfeng DING, Lin CHEN. Function Analysis of Sucrose Transporter OsSUT4 in Sucrose Transport in Rice [J]. Chinese Journal OF Rice Science, 2020, 34(6): 491-498. |
| [15] | Licheng ZHANG, Yixing LI, Tiankang WANG, Mudan QIU, Shufeng SONG, Hao DONG, Lei LI, Jianfeng LIU, Li LI. A Preliminary Study on the Function of Rice Heading Date Gene OsDof6 [J]. Chinese Journal OF Rice Science, 2020, 34(5): 397-405. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||