
Chinese Journal OF Rice Science ›› 2016, Vol. 30 ›› Issue (5): 469-478.DOI: 10.16819/j.1001-7216.2016.6009
• Orginal Article • Previous Articles Next Articles
Fang-quan WANG1,2, Fang-jun FAN1,2, Wen-qi LI1,2, Jin-yan ZHU1,2, Jun WANG1,2, Wei-gong ZHONG1,2, Jie YANG1,2,*( )
)
Received:2016-01-18
Revised:2016-03-03
Online:2016-09-10
Published:2016-09-10
Contact:
Jie YANG
王芳权1,2, 范方军1,2, 李文奇1,2, 朱金燕1,2, 王军1,2, 仲维功1,2, 杨杰1,2,*( )
)
通讯作者:
杨杰
基金资助:CLC Number:
Fang-quan WANG, Fang-jun FAN, Wen-qi LI, Jin-yan ZHU, Jun WANG, Wei-gong ZHONG, Jie YANG. Knock-out Efficiency Analysis of Pi21 Gene Using CRISPR/Cas9 in Rice[J]. Chinese Journal OF Rice Science, 2016, 30(5): 469-478.
王芳权, 范方军, 李文奇, 朱金燕, 王军, 仲维功, 杨杰. 利用CRISPR/Cas9技术敲除水稻Pi21基因的效率分析[J]. 中国水稻科学, 2016, 30(5): 469-478.
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.ricesci.cn/EN/10.16819/j.1001-7216.2016.6009
| 引物名称 Primer name | 引物序列(5'→3') Primer sequence (5'→3') |
|---|---|
| Pi21g-F | ACCAAAGCCTGTCTATCTGC |
| Pi21g-R | CACATCGATCAGCCTCGTG |
| Pi21UT1-F | GGCAGAAGCTGTGCAAGAAGATC |
| Pi21UT1-R | AAACGATCTTCTTGCACAGCTTC |
| Pi21UT2-F | GCCGCCGCTGCGATGCCAAGATC |
| Pi21UT2-R | AAACGATCTTGGCATCGCAGCGG |
| pU-F | CTCCGTTTTACCTGTGGAATCG |
| gRNA-R | CGGAGGAAAATTCCATCCAC |
| Pi21T1L-F | TTCAGAGGTCTCTCTCGCACTGGAATCGGCAGCAAAGG |
| Pi21T1L-R | AGCGTGGGTCTCGTCAGGGTCCATCCACTCCAAGCTC |
| Pi21T2L-F | TTCAGAGGTCTCTCTGACACTGGAATCGGCAGCAAAGG |
| Pi21T2L-R | AGCGTGGGTCTCGACCGGGTCCATCCACTCCAAGCTC |
| SP1 | CCCGACATAGATGCAATAACTTC |
| SP2 | GCGCGGTGTCATCTATGTTA |
| Pi21T1-F | AGGCTAATCAGCAGTGTTCCT |
| Pi21T1-R | CAGCTTGCACTCCGGCTTCG |
| Pi21T2-F | ATTGGTAACATTCGGCAAATT |
| Pi21T2-R | GTTCTTCACGTCGTACTCCA |
Table 1 Primers used in this research.
| 引物名称 Primer name | 引物序列(5'→3') Primer sequence (5'→3') |
|---|---|
| Pi21g-F | ACCAAAGCCTGTCTATCTGC |
| Pi21g-R | CACATCGATCAGCCTCGTG |
| Pi21UT1-F | GGCAGAAGCTGTGCAAGAAGATC |
| Pi21UT1-R | AAACGATCTTCTTGCACAGCTTC |
| Pi21UT2-F | GCCGCCGCTGCGATGCCAAGATC |
| Pi21UT2-R | AAACGATCTTGGCATCGCAGCGG |
| pU-F | CTCCGTTTTACCTGTGGAATCG |
| gRNA-R | CGGAGGAAAATTCCATCCAC |
| Pi21T1L-F | TTCAGAGGTCTCTCTCGCACTGGAATCGGCAGCAAAGG |
| Pi21T1L-R | AGCGTGGGTCTCGTCAGGGTCCATCCACTCCAAGCTC |
| Pi21T2L-F | TTCAGAGGTCTCTCTGACACTGGAATCGGCAGCAAAGG |
| Pi21T2L-R | AGCGTGGGTCTCGACCGGGTCCATCCACTCCAAGCTC |
| SP1 | CCCGACATAGATGCAATAACTTC |
| SP2 | GCGCGGTGTCATCTATGTTA |
| Pi21T1-F | AGGCTAATCAGCAGTGTTCCT |
| Pi21T1-R | CAGCTTGCACTCCGGCTTCG |
| Pi21T2-F | ATTGGTAACATTCGGCAAATT |
| Pi21T2-R | GTTCTTCACGTCGTACTCCA |

Fig. 1. Coding sequences co-linearity analysis of Pi21 gene. Nanjing 9108 is a blast sensitive variety, containing the susceptible allele Pi21. Owarihatamochi is a blast resistant variety, containing the resistant allele pi21.
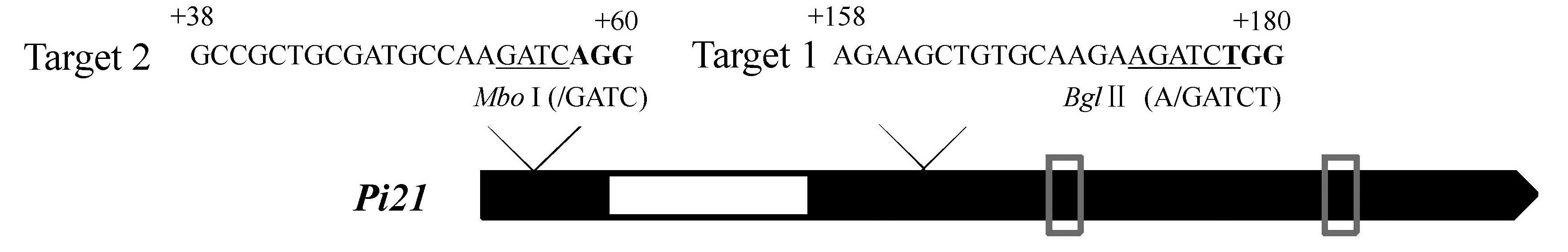
Fig. 2. Schematic diagram of Pi21 gene structure and gRNA targets. The base in bold indicates PAM; The restriction endonuclease recognized site is underlined; The black region indicates exon; The white region indicates intron; The gray box region indicates proline-rich motifs.
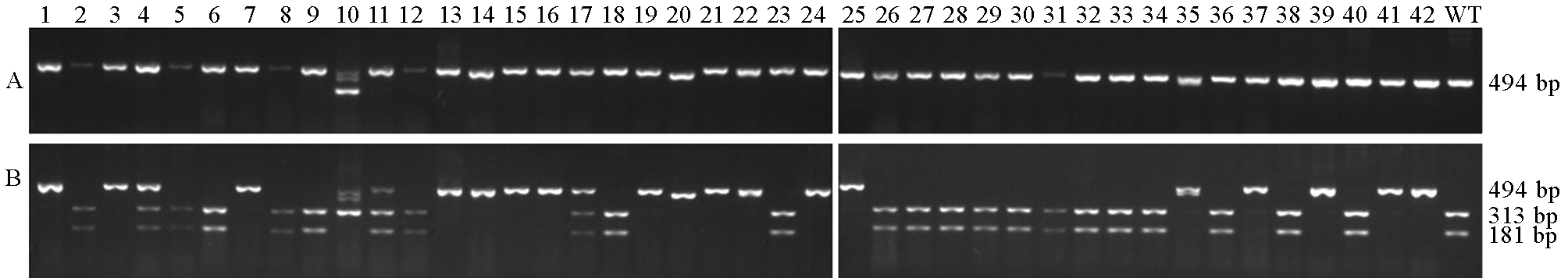
Fig. 5. Mutation identification at Target 1 by enzyme digestion. A, Before digestion; B, After digestion. 1~42, T0 transgenic lines; WT, Nanjing 9108 (wild-type).
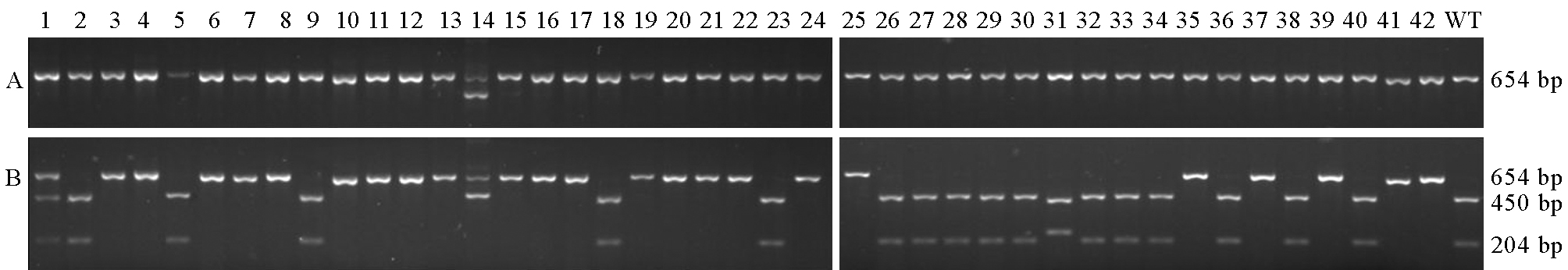
Fig. 6. Mutation identification at Target 2 by enzyme digestion. A, Before digestion; B, After digestion. 1~42, T0 transgenic lines; WT, Nanjing 9108 (wild-type).

Fig. 8. Analysis of mutation sequences of targets. “-1” and “-2” indicate different mutation type; The numbers in brackets are the numbers of sequenced clone; The arrows indicate the digestion sites of Cas9; The base in bold indicates protospacer adjacent motif(PAM); The base in red is the target sequence; The base in blue is the insert base; WT, Wild-type; #, Transgenic lines; d, Deletion; i, Insert.
| [1] | Belhaj K, Chaparro-Garcia A, Kamoun S, et al.Editing plant genomes with CRISPR/Cas9.Curr Opin Biotech, 2015, 32: 76-84. |
| [2] | Baltes N J, Voytas D F.Enabling plant synthetic biology through genome engineering.Trends Biotechnol, 2015, 33(2): 120-131. |
| [3] | 解莉楠, 宋凤艳, 张旸. CRISPR/Cas9 系统在植物基因组定点编辑中的研究进展. 中国农业科学, 2015, 48(9): 1669-1677. |
| Xie L N, Song F Y, Zhang Y.Progress in research of CRISPR/Cas9 system in genome targeted editing in plants.Sci Agric Sin, 2015, 48(9): 1669-1677. (in Chinese with English abstract) | |
| [4] | 李君, 张毅, 陈坤玲, 等. CRISPR/Cas 系统: RNA 靶向的基因组定向. 遗传, 2013, 35(11): 1265-1273. |
| Li J, Zhang Y, Chen K L, et al.CRISPR/Cas: A novel way of RNA-guided genome editing.Hereditas, 2013, 35(11): 1265-1273. (in Chinese with English abstract) | |
| [5] | Jiang W, Bikard D, Cox D, et al.RNA-guided editing of bacterial genomes using CRISPR-Cas systems.Nat Biotechnol, 2013, 31(3): 233-239. |
| [6] | DiCarlo J E, Norville J E, Mali P, et al. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems.Nucleic Acids Res, 2013, 41(7): 4336-4343. |
| [7] | Cong L, Ran F A, Cox D, et al.Multiplex genome engineering using CRISPR/Cas systems.Science, 2013, 339(6121): 819-823. |
| [8] | Mali P, Yang L, Esvelt K M, et al.RNA-guided human genome engineering via Cas9.Science, 2013, 339(6121): 823-826. |
| [9] | Wang H, Yang H, Shivalila C S, et al.One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering.Cell, 2013, 153(4): 910-918. |
| [10] | Feng Z, Zhang B, Ding W, et al.Efficient genome editing in plants using a CRISPR/Cas system.Cell Res, 2013, 23(10): 1229-1232. |
| [11] | Jiang W, Zhou H, Bi H, et al.Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice.Nucleic Acids Res, 2013, 41(20): e188. |
| [12] | Mao Y, Zhang H, Xu N, et al.Application of the CRISPR-Cas system for efficient genome engineering in plants.Mol Plant, 2013, 6(6): 2008-2011. |
| [13] | Shan Q, Wang Y, Li J, et al.Targeted genome modification of crop plants using a CRISPR-Cas system.Nat Biotechnol, 2013, 31(8): 686-688. |
| [14] | Zhang H, Zhang J, Wei P, et al.The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation.Plant Biotechnol J, 2014, 12(6): 797-807. |
| [15] | Li J F, Norville J E, Aach J, et al.Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9.Nat Biotechnol, 2013, 31(8): 688-691. |
| [16] | Nekrasov V, Staskawicz B, Weigel D, et al.Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease.Nat Biotechnol, 2013, 31(8): 691-693. |
| [17] | Liang Z, Zhang K, Chen K, et al.Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system.J Genet Genom, 2014, 41(2): 63-68. |
| [18] | Wang Y, Cheng X, Shan Q, et al.Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew.Nat Biotechnol, 2014, 32(9): 947-951. |
| [19] | Jacobs T B, LaFayette P R, Schmitz R J, et al. Targeted genome modifications in soybean with CRISPR/Cas9.BMC Biotechnol, 2015, 15(1): 16. |
| [20] | Li Z, Liu Z B, Xing A, et al.Cas9-guide RNA directed genome editing in soybean.Plant Physiol, 2015, 169(2): 960-970. |
| [21] | Sun X, Hu Z, Chen R, et al.Targeted mutagenesis in soybean using the CRISPR-Cas9 system.Sci Rep-UK, 2015, 5: 10342. |
| [22] | Brooks C, Nekrasov V, Lippman Z B, et al.Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system.Plant Physiol, 2014, 166(3): 1292-1297. |
| [23] | Miao J, Guo D, Zhang J, et al.Targeted mutagenesis in rice using CRISPR-Cas system.Cell Res, 2013, 23(10): 1233-1236. |
| [24] | Shan Q, Wang Y, Li J, et al.Genome editing in rice and wheat using the CRISPR/Cas system.Nat Protec, 2014, 9(10): 2395-2410. |
| [25] | Xu R F, Li H, Qin R Y, et al.Generation of inheritable and “transgene clean” targeted genome-modified rice in later generations using the CRISPR/Cas9 system.Sci Rep-UK, 2015, 5: 11491. |
| [26] | Ma X, Zhang Q, Zhu Q, et al.A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants.Mol Plant, 2015, 8(8): 1274-1284. |
| [27] | 鄂志国, 王磊. 水稻抗病性基因的克隆和功能研究进展. 遗传, 2009, 31(10): 999-1005. |
| E Z G, Wang L. Advance on the cloning and functional analysis of disease resistance genes in rice.Hereditas, 2009, 31(10): 999-1005. (in Chinese with English abstract) | |
| [28] | Fukuoka S, Okuno K.QTL analysis and mapping of pi21, a recessive gene for field resistance to rice blast in Japanese upland rice.Theor Appl Genet, 2001, 103(2): 185-190. |
| [29] | Fukuoka S, Saka N, Koga H, et al.Loss of function of a proline-containing protein confers durable disease resistance in rice.Science, 2009, 325(5943): 998-1001. |
| [30] | 朱金燕, 王军, 范方军, 等. 水稻稻瘟病广谱抗病新等位基因pi21t的鉴定及其抗性应用. 华北农学报, 2014, 29(6): 11-15. |
| Zhu J Y, Wang J, Fan F J.et al.Identification and application of one new rice blast broad-spectrum resistance allele pi21t.Acta Agric Bor-Sin, 2014, 29(6): 11-15. (in Chinese with English abstract) | |
| [31] | 王才林, 张亚东, 朱镇, 等. 优良食味粳稻新品种南粳9108的选育与利用. 江苏农业科学, 2013, 41(9): 86-88. |
| Wang C L, Zhang Y D, Zhu Z, et al.Breeding and application of Nanjing 9108, a new japonica variety with excellent eating quality.Jiangsu Agric Sci, 2013, 41(9): 86-88. | |
| [32] | Xie K, Yang Y.RNA-guided genome editing in plants using a CRISPR-Cas system.Mol Plant, 2013, 6(6): 1975-1983. |
| [33] | 朱立宏. 关于我国水稻高产育种的我见. 南京农业大学学报, 2007, 30(1): 129-135. |
| Zhu L H.Some critical considerations on rice high-yielding breeding in China.J Nanjing Agric Univ, 2007, 30(1): 129-135. (in Chinese with English abstract) | |
| [34] | 凌忠专, 雷财林, 王久林. 稻瘟病菌生理小种研究的回顾与展望. 中国农业科学, 2004, 37(12): 1849-1859. |
| Ling Z Z, Lei C L, Wang J L.Review and prospect for study of physiologic races on rice blast fungus (Pyricularia grasea).Sci Agric Sin, 2004, 37(12): 1849-1859. (in Chinese with English abstract) |
| [1] | XU Danjie, LIN Qiaoxia, LI Zhengkang, ZHUANG Xiaoqian, LING Yu, LAI Meiling, CHEN Xiaoting, LU Guodong. OsOPR10 Positively Regulates Rice Blast and Bacterial Blight Resistance [J]. Chinese Journal OF Rice Science, 2024, 38(4): 364-374. |
| [2] | CHEN Mingliang, XIONG Wentao, SHEN Yumin, XIONG Huanjin, LUO Shiyou, WU Xiaoyan, HU Lanxiang, XIAO Yeqing. Genetic Dissection of Broad Spectrum Resistance of the Rice Maintainer Ganxiang B [J]. Chinese Journal OF Rice Science, 2023, 37(5): 470-477. |
| [3] | LI Jingfang, WEN Shuyue, ZHAO Lijun, CHEN Tingmu, ZHOU Zhenling, SUN Zhiguang, LIU Yan, CHEN Haiyuan, ZHANG Yunhui, CHI Ming, XING Yungao, XU Bo, XU Dayong, WANG Baoxiang. Development of Aromatic Salt-tolerant Rice Based on CRISPR/Cas9 Technology [J]. Chinese Journal OF Rice Science, 2023, 37(5): 478-485. |
| [4] | LI Gang, GAO Qingsong, LI Wei, ZHANG Wenxia, WANG Jian, CHEN Baoshan, WANG Di, GAO Hao, XU Weijun, CHEN Hongqi, JI Jianhui. Directed Knockout of SD1 Gene Improves Lodging Resistance and Blast Resistance of Rice [J]. Chinese Journal OF Rice Science, 2023, 37(4): 359-367. |
| [5] | DUAN Min, XIE Liujie, GAO Xiuying, TANG Haijuan, HUANG Shanjun, PAN Xiaobiao. Creation of Thermo-sensitive Genic Male Sterile Rice Lines with Wide Compatibility Based on CRISPR/Cas9 Technology [J]. Chinese Journal OF Rice Science, 2023, 37(3): 233-243. |
| [6] | WANG Shiguang, LU Zhanhua, LIU Wei, LU Dongbai, WANG Xiaofei, FANG Zhiqiang, WU Haoxiang, HE Xiuying. Generating Guangdong Simiao Rice Germplasms by Applying CRISPR/Cas9 Gene Editing and Marker-assisted Selection Technology [J]. Chinese Journal OF Rice Science, 2023, 37(1): 29-36. |
| [7] | ZHANG Yuanye, YIN Liying, LI Rongtian, HE Mingliang, LIU Xinxin, PAN Tingting, TIAN Xiaojie, BU Qingyun, LI Xiufeng. Breeding of Rc Function Restoration Red Rice via CRISPR/Cas9 Mediated Genome Editing [J]. Chinese Journal OF Rice Science, 2022, 36(6): 572-578. |
| [8] | YIN Liying, ZHANG Yuanye, LI Rongtian, HE Mingliang, WANG Fangquan, XU Yang, LIU Xinxin, PAN Tingting, TIAN Xiaojie, BU Qingyun, LI Xiufeng. Improvement of Herbicide Resistance in Rice by Using CRISPR/Cas9 System [J]. Chinese Journal OF Rice Science, 2022, 36(5): 459-466. |
| [9] | ZHOU Yonglin, SHEN Xiaolei, ZHOU Lishuai, LIN Qiaoxia, WANG Zhaolu, CHEN Jing, FENG Huijie, ZHANG Zhenwen, CHEN Xiaoting, LU Guodong. OsLOX10 Positively Regulates Defense Responses of Rice to Rice Blast and Bacterial Blight [J]. Chinese Journal OF Rice Science, 2022, 36(4): 348-356. |
| [10] | LI Zhaowei, SUN Congying, LING Donglan, ZENG Huiling, ZHANG Xiaomei, FAN Kai, LIN Wenxiong. Construction of osarf7 Mutants in Rice Based on CRISPR/Cas9 Technology and Investigation on Their Agronomic Traits [J]. Chinese Journal OF Rice Science, 2022, 36(3): 237-247. |
| [11] | LIANG Minmin, ZHANG Huali, CHEN Junyu, DAI Dongqing, DU Chengxing, WANG Huimei, MA Liangyong. Developing Fragrant Early indica TGMS Line with Blast Resistance by Using CRISPR/Cas9 Technology [J]. Chinese Journal OF Rice Science, 2022, 36(3): 248-258. |
| [12] | Yudong CAO, Xiangyi XIAO, Naizhong YE, Xiaowen DING, Xiaoxuan YI, Jinling LIU, Yinghui XIAO. Auxin Regulator OsGRF4 Simultaneously Regulates Rice Grain Shape and Blast Resistance [J]. Chinese Journal OF Rice Science, 2021, 35(6): 629-638. |
| [13] | Kai LU, Tao CHEN, Shu YAO, Wenhua LIANG, Xiaodong WEI, Yadong ZHANG, Cailin WANG. Functional Analysis on Four Receptor-like Protein Kinases Under Salt Stress in Rice [J]. Chinese Journal OF Rice Science, 2021, 35(2): 103-111. |
| [14] | Tianshun ZHOU, Dong YU, Ling LIU, Ning OUYANG, Guilong YUAN, Meijuan DUAN, Dingyang YUAN. CRISPR/Cas9-mediatedEditing of AFP1Improves Rice Stress Tolerance [J]. Chinese Journal OF Rice Science, 2021, 35(1): 11-18. |
| [15] | Mengzhu LI, Gaopeng WANG, Yue WU, Yi REN, Ganghua LI, Zhenghui LIU, Yanfeng DING, Lin CHEN. Function Analysis of Sucrose Transporter OsSUT4 in Sucrose Transport in Rice [J]. Chinese Journal OF Rice Science, 2020, 34(6): 491-498. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||