
中国水稻科学 ›› 2025, Vol. 39 ›› Issue (6): 789-800.DOI: 10.16819/j.1001-7216.2025.240807
王世林1, 吴婷1, 周诗琪1, 宋思铭2, 胡标林1,*( )
)
收稿日期:2024-08-07
修回日期:2024-10-14
出版日期:2025-11-10
发布日期:2025-11-19
通讯作者:
* email:hubiaolin992@126.com
基金资助:
WANG Shilin1, WU Ting1, ZHOU Shiqi1, SONG Siming2, HU Biaolin1,*( )
)
Received:2024-08-07
Revised:2024-10-14
Online:2025-11-10
Published:2025-11-19
Contact:
* email:hubiaolin992@126.com
摘要:
【目的】水稻种子储藏对保障粮食安全生产具有重要意义,挖掘水稻耐储性基因资源,为培育耐储藏水稻新品种奠定遗传基础。【方法】以东乡野生稻东野80为供体、栽培稻R974为轮回亲本,分别构建了364份回交重组自交系和84份BC3F7单片段代换系。统计人工老化处理种子发芽率,并结合QTL和BSA-seq分析联合检测水稻耐储性QTL,进一步通过基因注释信息、GO富集分析和基因测序预测候选基因。【结果】利用单片段代换系共检测到9个耐储性QTL,分别为qSS1.1、qSS1.2、qSS2.1、qSS2.2、qSS3、qSS6、qSS7、qSS9.1和qSS11,贡献率为1.60%~17.82%,其中7个QTL的增效等位基因来自东野80,且qSS7和qSS11为新的QTL。而利用回交重组自交系进行BSA-seq分析,共关联到37个位点,分布于2、4、5、6、7、8、9和11号染色体上。其中,qSS7在2种方法中均被检测,将其进一步缩小至480.1 kb区间内。此外,qSS4和qSS9.2分别被缩小至199.7 kb和177.8 kb的交叠区间内。结合基因注释信息查询、GO富集分析和基因测序结果,推测LOC_Os04g31040和LOC_Os04g31070为qSS4的候选基因,LOC_Os07g07930为qSS7的候选基因,LOC_Os09g15800和LOC_Os09g15835为qSS9.2的候选基因。【结论】本研究通过QTL和BSA-Seq分析分别鉴定到9个和37个耐储性QTL,并推测了qSS4、qSS7和qSS9.2的候选基因。其中,qSS4和qSS9.2的东野80增效等位基因显著提高水稻种子耐储性,可为东乡野生稻耐储性QTL克隆及其在栽培稻耐储性遗传育种利用提供新的有利位点。
王世林, 吴婷, 周诗琪, 宋思铭, 胡标林. 结合BSA-seq和QTL分析鉴定东乡野生稻耐储性QTL[J]. 中国水稻科学, 2025, 39(6): 789-800.
WANG Shilin, WU Ting, ZHOU Shiqi, SONG Siming, HU Biaolin. Identification of QTLs for Seed Storability in Dongxiang Wild Rice by Integrating BSA-Seq and QTL Analysis[J]. Chinese Journal OF Rice Science, 2025, 39(6): 789-800.
| 名称 Name | 正向引物 Forward primer (5'-3') | 反向引物 Reverse primer (5'-3') | 候选基因 Candidate gene |
|---|---|---|---|
| Fou31040 | CAGGTCAGAGGAACTCAG | GGATGCATTAGCAGATTC | LOC_Os04g31040 |
| Fou31070 | GATCTCGATGAGGAGGTG | GCAGATGGACAGGTAATTG | LOC_Os04g31070 |
| Fou31240 | GCGATATCCTATAGCCAA | CAGTCTAGCAATTCATGC | LOC_Os04g31240 |
| Sev07420 | GTGCTCGATGTGGCTTAG | CATGCATGTTGCAATGC | LOC_Os07g07420 |
| Sev07790 | CAATCACCTCATCACACAC | CATGGCACACTCAACAC | LOC_Os07g07790 |
| Sev07860 | TGAGACCAATGGAAATGG | GATTCCATCACTTCACAC | LOC_Os07g07860 |
| Sev07870 | CTGAGAGACATACATCAATC | GATACTCATCAGAAGGCATAC | LOC_Os07g07870 |
| Sev07880 | CTTTACCGAATGGAGACCAG | GGATTTACACTAGCAAGT | LOC_Os07g07880 |
| Sev07930 | CAGACACCAAGAGCATC | CGAATTCCTACACGCAC | LOC_Os07g07930 |
| Nin15800 | GACCGAGTGAGTCATCAAG | GTGAGTACTCTACATGATAC | LOC_Os09g15800 |
| Nin15810 | CACAGTATCTTCCTGCTAC | CAAGCATGGACATGCTAATC | LOC_Os09g15810 |
| Nin15835 | CTCTGGTTAGATCTTGCAC | CAATATGAATACCCTGCA | LOC_Os09g15835 |
表1 扩增候选基因的引物信息
Table 1. Primer information of amplified candidate genes
| 名称 Name | 正向引物 Forward primer (5'-3') | 反向引物 Reverse primer (5'-3') | 候选基因 Candidate gene |
|---|---|---|---|
| Fou31040 | CAGGTCAGAGGAACTCAG | GGATGCATTAGCAGATTC | LOC_Os04g31040 |
| Fou31070 | GATCTCGATGAGGAGGTG | GCAGATGGACAGGTAATTG | LOC_Os04g31070 |
| Fou31240 | GCGATATCCTATAGCCAA | CAGTCTAGCAATTCATGC | LOC_Os04g31240 |
| Sev07420 | GTGCTCGATGTGGCTTAG | CATGCATGTTGCAATGC | LOC_Os07g07420 |
| Sev07790 | CAATCACCTCATCACACAC | CATGGCACACTCAACAC | LOC_Os07g07790 |
| Sev07860 | TGAGACCAATGGAAATGG | GATTCCATCACTTCACAC | LOC_Os07g07860 |
| Sev07870 | CTGAGAGACATACATCAATC | GATACTCATCAGAAGGCATAC | LOC_Os07g07870 |
| Sev07880 | CTTTACCGAATGGAGACCAG | GGATTTACACTAGCAAGT | LOC_Os07g07880 |
| Sev07930 | CAGACACCAAGAGCATC | CGAATTCCTACACGCAC | LOC_Os07g07930 |
| Nin15800 | GACCGAGTGAGTCATCAAG | GTGAGTACTCTACATGATAC | LOC_Os09g15800 |
| Nin15810 | CACAGTATCTTCCTGCTAC | CAAGCATGGACATGCTAATC | LOC_Os09g15810 |
| Nin15835 | CTCTGGTTAGATCTTGCAC | CAATATGAATACCCTGCA | LOC_Os09g15835 |
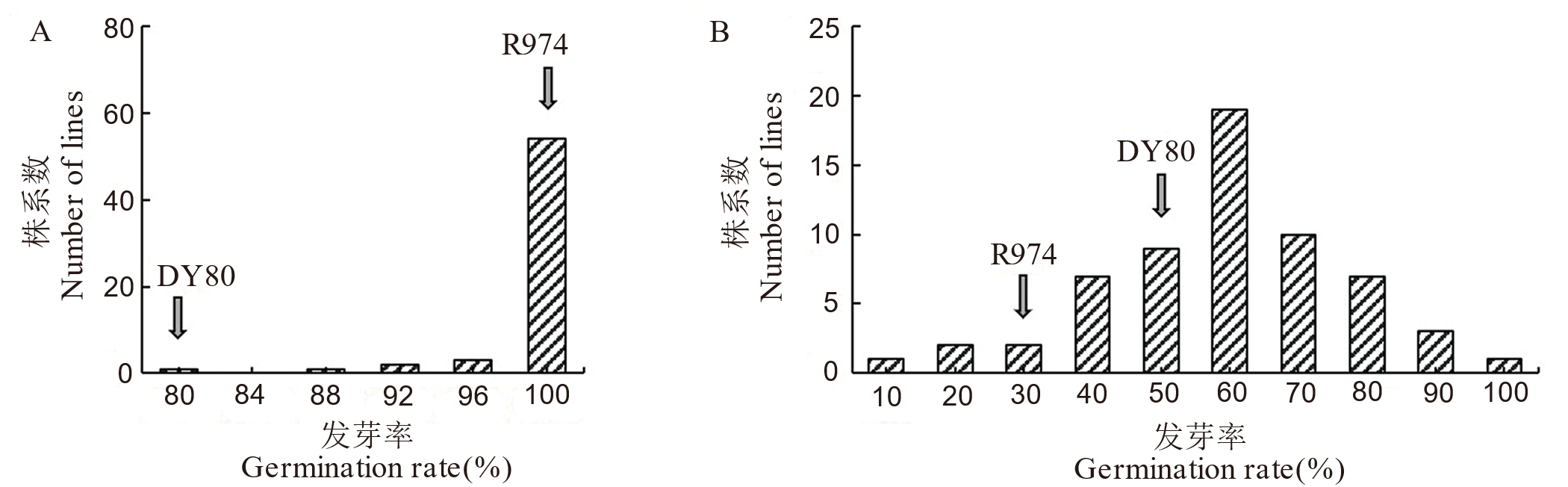
图1 BG群体的表型分布 A:正常萌发试验的表型分布;B:人工陈化处理后的表型分布。
Fig.1. Phenotypic distribution in BG population A, Distribution of germination rate in natural conditions; B, Distribution of germination rate after artificial aging treatment.
| 染色体 Chromosome | QTL | 区间 Interval | 位置 Position | LOD值 LOD | 加性效应 A | 贡献率 R2(%) | 已克隆基因 Cloned genes |
|---|---|---|---|---|---|---|---|
| 1 | qSS1.1 | chr1-204.9-chr1-216.3 | 36,735,178-40,016,503 | 2.55 | 13.77 | 1.64 | |
| 1 | qSS1.2 | chr1-216.3-chr1-230.0 | 40,016,503-41,198,359 | 3.07 | 9.19 | 2.23 | |
| 2 | qSS2.1 | chr2-84.6-chr2-101.9 | 27,538,266-28,417,539 | 6.77 | 13.54 | 4.78 | |
| 2 | qSS2.2 | chr2-101.9-chr2-123.4 | 28,417,539-33,682,808 | 7.97 | 13.40 | 4.79 | OsbZIP23 |
| 3 | qSS3 | chr3-111.7-chr3-121.4 | 30,677,202-33,003,423 | 2.56 | 14.09 | 1.60 | OsLOX2 |
| 6 | qSS6 | chr6-34.7-chr6-50.0 | 17,953,681-24,234,137 | 10.29 | −24.17 | 7.70 | |
| 7 | qSS7 | chr7-21.1-chr7-25.2 | 3,146,214-5,200,751 | 8.82 | −30.51 | 6.24 | |
| 9 | qSS9.1 | chr9-69.2-chr9-78.9 | 13,164,222-14,567,027 | 3.33 | 10.01 | 2.09 | |
| 11 | qSS11 | chr11-37.6-chr11-48.2 | 8,990,534-9,368,443 | 17.36 | 23.81 | 17.82 |
表2 BG群体检测到的耐储性QTL
Table 2. Detected QTLs for seed storability in BG population
| 染色体 Chromosome | QTL | 区间 Interval | 位置 Position | LOD值 LOD | 加性效应 A | 贡献率 R2(%) | 已克隆基因 Cloned genes |
|---|---|---|---|---|---|---|---|
| 1 | qSS1.1 | chr1-204.9-chr1-216.3 | 36,735,178-40,016,503 | 2.55 | 13.77 | 1.64 | |
| 1 | qSS1.2 | chr1-216.3-chr1-230.0 | 40,016,503-41,198,359 | 3.07 | 9.19 | 2.23 | |
| 2 | qSS2.1 | chr2-84.6-chr2-101.9 | 27,538,266-28,417,539 | 6.77 | 13.54 | 4.78 | |
| 2 | qSS2.2 | chr2-101.9-chr2-123.4 | 28,417,539-33,682,808 | 7.97 | 13.40 | 4.79 | OsbZIP23 |
| 3 | qSS3 | chr3-111.7-chr3-121.4 | 30,677,202-33,003,423 | 2.56 | 14.09 | 1.60 | OsLOX2 |
| 6 | qSS6 | chr6-34.7-chr6-50.0 | 17,953,681-24,234,137 | 10.29 | −24.17 | 7.70 | |
| 7 | qSS7 | chr7-21.1-chr7-25.2 | 3,146,214-5,200,751 | 8.82 | −30.51 | 6.24 | |
| 9 | qSS9.1 | chr9-69.2-chr9-78.9 | 13,164,222-14,567,027 | 3.33 | 10.01 | 2.09 | |
| 11 | qSS11 | chr11-37.6-chr11-48.2 | 8,990,534-9,368,443 | 17.36 | 23.81 | 17.82 |
| 样品 Sample | 原始数据 Raw base (bp) | 过滤后的有效数据 Clean base (bp) | 比对率 Mapping ratio (%) | 平均测序深度 Average depth (×) | 大于或等于30的碱基所占百分比 Clean Q30 (%) |
|---|---|---|---|---|---|
| R974 | 11,531,262,900 | 10,612,006,654 | 99.04 | 30.41 | 88.59 |
| 东野80 Dongye 80 | 12,730,838,700 | 12,019,467,219 | 98.55 | 32.58 | 94.20 |
| S池S-bulk | 11,921,866,200 | 11,435,986,027 | 97.44 | 31.16 | 92.52 |
| W池W-bulk | 11,710,934,400 | 11,116,889,747 | 97.73 | 30.00 | 93.70 |
表3 用于BSA分析的测序数据概况
Table 3. Summary of sequencing data for BSA-seq analysis
| 样品 Sample | 原始数据 Raw base (bp) | 过滤后的有效数据 Clean base (bp) | 比对率 Mapping ratio (%) | 平均测序深度 Average depth (×) | 大于或等于30的碱基所占百分比 Clean Q30 (%) |
|---|---|---|---|---|---|
| R974 | 11,531,262,900 | 10,612,006,654 | 99.04 | 30.41 | 88.59 |
| 东野80 Dongye 80 | 12,730,838,700 | 12,019,467,219 | 98.55 | 32.58 | 94.20 |
| S池S-bulk | 11,921,866,200 | 11,435,986,027 | 97.44 | 31.16 | 92.52 |
| W池W-bulk | 11,710,934,400 | 11,116,889,747 | 97.73 | 30.00 | 93.70 |
| 染色体 Chromosome | 起始位置 Start position (bp) | 终止位置 Stop position (bp) | 长度 Length (bp) | 基因数量 Gene number | 已克隆基因 Cloned gene |
|---|---|---|---|---|---|
| 2 | 23,101,936 | 23,699,539 | 597,604 | 89 | |
| 2 | 24,250,015 | 24,598,471 | 348,457 | 51 | |
| 2 | 25,600,427 | 25,999,871 | 399,445 | 64 | |
| 4 | 2,500,027 | 3,032,263 | 532,237 | 17 | |
| 4 | 3,377,104 | 3,848,072 | 470,969 | 26 | |
| 4 | 4,650,046 | 4,697,640 | 47,595 | 4 | |
| 4 | 6,202,257 | 6,499,011 | 296,755 | 20 | |
| 4 | 6,557,412 | 6,599,738 | 42,327 | 4 | |
| 4 | 12,653,023 | 12,699,529 | 46,507 | 3 | |
| 4 | 17,550,918 | 17,973,774 | 422,857 | 20 | |
| 4 | 18,501,088 | 18,749,740 | 248,653 | 33 | |
| 5 | 2,360,354 | 2,498,194 | 137,841 | 18 | |
| 5 | 2,834,017 | 3,530,935 | 696,919 | 104 | |
| 5 | 6,000,030 | 6,433,133 | 433,104 | 36 | |
| 5 | 12,220,484 | 12,675,873 | 455,390 | 12 | |
| 5 | 16,703,352 | 16,798,240 | 94,889 | 6 | |
| 6 | 3,956,957 | 3,993,286 | 36,330 | 10 | |
| 6 | 4,308,227 | 4,920,955 | 612,729 | 101 | |
| 6 | 5,108,294 | 5,390,250 | 281,957 | 38 | OsbZIP46 |
| 7 | 450,272 | 547,103 | 96,832 | 16 | |
| 7 | 3,657,270 | 4,137,319 | 480,050 | 75 | |
| 7 | 25,802,892 | 25,965,970 | 163,079 | 22 | |
| 7 | 26,080,904 | 26,992,642 | 911,739 | 153 | OsPER1A |
| 7 | 27,404,044 | 28,395,518 | 991,475 | 183 | |
| 7 | 28,650,196 | 29,672,511 | 1,022,316 | 193 | |
| 8 | 2,500,187 | 3,047,703 | 547,517 | 41 | |
| 8 | 24,509,072 | 24,645,822 | 136,751 | 21 | |
| 8 | 24,702,479 | 24,749,260 | 46,782 | 11 | |
| 9 | 9,602,172 | 9,849,880 | 247,709 | 27 | |
| 9 | 9,910,243 | 10,029,893 | 119,651 | 9 | |
| 9 | 11,421,565 | 11,635,435 | 213,871 | 14 | |
| 11 | 13,750,043 | 14,348,927 | 598,885 | 49 | |
| 11 | 16,650,014 | 17,099,579 | 449,566 | 36 | |
| 11 | 20,419,176 | 20,649,984 | 230,809 | 23 | |
| 11 | 20,750,146 | 20,799,935 | 49,790 | 5 | |
| 11 | 22,500,104 | 22,599,573 | 99,470 | 18 | |
| 11 | 22,650,021 | 22,799,987 | 149,967 | 14 |
表4 BSA关联的候选区间
Table 4. Candidate regions detected by BSA-seq analysis
| 染色体 Chromosome | 起始位置 Start position (bp) | 终止位置 Stop position (bp) | 长度 Length (bp) | 基因数量 Gene number | 已克隆基因 Cloned gene |
|---|---|---|---|---|---|
| 2 | 23,101,936 | 23,699,539 | 597,604 | 89 | |
| 2 | 24,250,015 | 24,598,471 | 348,457 | 51 | |
| 2 | 25,600,427 | 25,999,871 | 399,445 | 64 | |
| 4 | 2,500,027 | 3,032,263 | 532,237 | 17 | |
| 4 | 3,377,104 | 3,848,072 | 470,969 | 26 | |
| 4 | 4,650,046 | 4,697,640 | 47,595 | 4 | |
| 4 | 6,202,257 | 6,499,011 | 296,755 | 20 | |
| 4 | 6,557,412 | 6,599,738 | 42,327 | 4 | |
| 4 | 12,653,023 | 12,699,529 | 46,507 | 3 | |
| 4 | 17,550,918 | 17,973,774 | 422,857 | 20 | |
| 4 | 18,501,088 | 18,749,740 | 248,653 | 33 | |
| 5 | 2,360,354 | 2,498,194 | 137,841 | 18 | |
| 5 | 2,834,017 | 3,530,935 | 696,919 | 104 | |
| 5 | 6,000,030 | 6,433,133 | 433,104 | 36 | |
| 5 | 12,220,484 | 12,675,873 | 455,390 | 12 | |
| 5 | 16,703,352 | 16,798,240 | 94,889 | 6 | |
| 6 | 3,956,957 | 3,993,286 | 36,330 | 10 | |
| 6 | 4,308,227 | 4,920,955 | 612,729 | 101 | |
| 6 | 5,108,294 | 5,390,250 | 281,957 | 38 | OsbZIP46 |
| 7 | 450,272 | 547,103 | 96,832 | 16 | |
| 7 | 3,657,270 | 4,137,319 | 480,050 | 75 | |
| 7 | 25,802,892 | 25,965,970 | 163,079 | 22 | |
| 7 | 26,080,904 | 26,992,642 | 911,739 | 153 | OsPER1A |
| 7 | 27,404,044 | 28,395,518 | 991,475 | 183 | |
| 7 | 28,650,196 | 29,672,511 | 1,022,316 | 193 | |
| 8 | 2,500,187 | 3,047,703 | 547,517 | 41 | |
| 8 | 24,509,072 | 24,645,822 | 136,751 | 21 | |
| 8 | 24,702,479 | 24,749,260 | 46,782 | 11 | |
| 9 | 9,602,172 | 9,849,880 | 247,709 | 27 | |
| 9 | 9,910,243 | 10,029,893 | 119,651 | 9 | |
| 9 | 11,421,565 | 11,635,435 | 213,871 | 14 | |
| 11 | 13,750,043 | 14,348,927 | 598,885 | 49 | |
| 11 | 16,650,014 | 17,099,579 | 449,566 | 36 | |
| 11 | 20,419,176 | 20,649,984 | 230,809 | 23 | |
| 11 | 20,750,146 | 20,799,935 | 49,790 | 5 | |
| 11 | 22,500,104 | 22,599,573 | 99,470 | 18 | |
| 11 | 22,650,021 | 22,799,987 | 149,967 | 14 |
| QTL | 候选基因 Candidate gene | 位置 Position(bp) | 注释信息 Annotation information | GO |
|---|---|---|---|---|
| qSS4 | LOC_Os04g31040 | 18,553,801−18,551,238 | 紫黄质脱环氧化酶 Violaxanthin de-epoxidase | 细胞脂质代谢过程 Cellular lipid metabolic process |
| LOC_Os04g31070 | 18,564,417−18,560,177 | 酰基去饱和酶 Acyl-desaturase | 氧化还原酶活性 Oxidoreductase activity | |
| LOC_Os04g31240 | 18,690,646−18,694,747 | 水解酶 Hydrolase | 脂质稳态 Lipid homeostasis | |
| qSS7 | LOC_Os07g07420 | 3,702,287−3,705,030 | 赤霉素20氧化酶1-B Gibberellin 20 oxidase 1-B | 赤霉素生物合成过程 Gibberellin biosynthetic process |
| LOC_Os07g07790 | 3,910,568−3,911,638 | 脂质转运蛋白家族蛋白前体75 LTPL75: Lipid transfer protein family protein precursor | 脂质结合 Lipid binding | |
| LOC_Os07g07860 | 3,949,371−3,947,968 | 脂质转运蛋白家族蛋白前体76 LTPL76: Lipid transfer protein family protein precursor | 脂质结合 Lipid binding | |
| LOC_Os07g07870 | 3,950,800−3,951,581 | 脂质转运蛋白家族蛋白前体77 LTPL77: Lipid transfer protein family protein precursor | 脂质结合 Lipid binding | |
| LOC_Os07g07880 | 3,954,894−3,952,981 | 色氨酸阻遏蛋白 Trp repressor | 组蛋白甲基转移酶复合体 Histone methyltransferase complex | |
| LOC_Os07g07930 | 3,980,211−3,981,284 | 脂质转运蛋白家族蛋白前体78 LTPL78: Lipid transfer protein family protein precursor | 脂质结合 Lipid binding | |
| qSS9.2 | LOC_Os09g15800 | 9,650,347−9,647,717 | 四肽样螺旋蛋白 Tetratricopeptide-like helical | 种子寿命的获得 Acquisition of seed longevity |
| LOC_Os09g15810 | 9,652,178−9,654,972 | 双功能蛋白folD Bifunctional protein folD | 氧化还原酶活性 Oxidoreductase activity | |
| LOC_Os09g15835 | 9,665,794−9,663,456 | OBP32pep | 茉莉酸生物合成过程 Jasmonic acid biosynthetic process |
表5 候选基因注释信息
Table 5. Annotation information of candidate genes
| QTL | 候选基因 Candidate gene | 位置 Position(bp) | 注释信息 Annotation information | GO |
|---|---|---|---|---|
| qSS4 | LOC_Os04g31040 | 18,553,801−18,551,238 | 紫黄质脱环氧化酶 Violaxanthin de-epoxidase | 细胞脂质代谢过程 Cellular lipid metabolic process |
| LOC_Os04g31070 | 18,564,417−18,560,177 | 酰基去饱和酶 Acyl-desaturase | 氧化还原酶活性 Oxidoreductase activity | |
| LOC_Os04g31240 | 18,690,646−18,694,747 | 水解酶 Hydrolase | 脂质稳态 Lipid homeostasis | |
| qSS7 | LOC_Os07g07420 | 3,702,287−3,705,030 | 赤霉素20氧化酶1-B Gibberellin 20 oxidase 1-B | 赤霉素生物合成过程 Gibberellin biosynthetic process |
| LOC_Os07g07790 | 3,910,568−3,911,638 | 脂质转运蛋白家族蛋白前体75 LTPL75: Lipid transfer protein family protein precursor | 脂质结合 Lipid binding | |
| LOC_Os07g07860 | 3,949,371−3,947,968 | 脂质转运蛋白家族蛋白前体76 LTPL76: Lipid transfer protein family protein precursor | 脂质结合 Lipid binding | |
| LOC_Os07g07870 | 3,950,800−3,951,581 | 脂质转运蛋白家族蛋白前体77 LTPL77: Lipid transfer protein family protein precursor | 脂质结合 Lipid binding | |
| LOC_Os07g07880 | 3,954,894−3,952,981 | 色氨酸阻遏蛋白 Trp repressor | 组蛋白甲基转移酶复合体 Histone methyltransferase complex | |
| LOC_Os07g07930 | 3,980,211−3,981,284 | 脂质转运蛋白家族蛋白前体78 LTPL78: Lipid transfer protein family protein precursor | 脂质结合 Lipid binding | |
| qSS9.2 | LOC_Os09g15800 | 9,650,347−9,647,717 | 四肽样螺旋蛋白 Tetratricopeptide-like helical | 种子寿命的获得 Acquisition of seed longevity |
| LOC_Os09g15810 | 9,652,178−9,654,972 | 双功能蛋白folD Bifunctional protein folD | 氧化还原酶活性 Oxidoreductase activity | |
| LOC_Os09g15835 | 9,665,794−9,663,456 | OBP32pep | 茉莉酸生物合成过程 Jasmonic acid biosynthetic process |

图4 注释基因的GO富集分析结果 A: qSS4候选区间内注释基因的GO富集分析; B: qSS7候选区间内注释基因的GO富集分析; C: qSS9.2候选区间内注释基因的GO富集分析。
Fig. 4. Results of GO enrichment analysis for annotated genes A, Results of GO enrichment analysis for annotated genes in the interval of qSS4; B, Results of GO enrichment analysis for annotated genes in the interval of qSS7; C, Results of GO enrichment analysis for annotated genes in the interval of qSS9.2.
| [1] | 黄祎雯, 孙滨, 程灿, 牛付安, 周继华, 张安鹏, 涂荣剑, 李瑶, 姚瑶. 对水稻种子耐储性QTL的研究[J]. 作物学报, 2022, 48(9): 2255-2264. |
| Huang Y W, Sun B, Cheng C, Niu F A, Zhou J H, Zhang A P, Tu R J, Li Y, Yao Y. QTL mapping of seed storage tolerance in rice[J]. Acta Agronomica Sinica, 2022, 48(9): 2255-2264. (in Chinese with English abstract) | |
| [2] | Xu H W, Zhu Y, Lian L, Zhang J. Antisense suppression of lox3 gene in endosperm enhances seed longevity[J]. Plant Biotechnology Journal, 2015, 13(4): 526-539. |
| [3] | 费月新, 曹玉洁, 吴敏, 吴洪恺. 稻米储藏品质劣变机制研究进展与展望[J]. 中国稻米, 2018, 24(5): 22-26. |
| Fei Y X, Cao Y J, Wu M, Wu H K. Research progress and prospect on the mechanism of rice grain storage quality deterioration[J]. China Rice, 2018, 24(5): 22-26. (in Chinese with English abstract) | |
| [4] | 黄上志, 傅家瑞. 贮藏温度和相对湿度对杂交水稻种子耐藏性影响[J]. 植物生理学通讯, 1986, 11(6): 38-41. |
| Huang S Z, Fu J R. Effect of storage temperature and relative humidity on the storability of hybrid rice seeds[J]. Plant Physiology Communications, 1986, 11(6): 38-41. (in Chinese with English abstract) | |
| [5] | 黄祎雯, 张维谊, 王霞, 王敏, 沈斯文, 高猛峰, 梅博, 汪弘康, 童金蓉, 曹黎明, 丰东升, 孙滨. 33份不同基因型水稻耐储性比较[J/OL]. 分子植物育种, 2024, https://link.cnki.net/urlid/46.1068.S.20240202.1646.010. |
| Huang Y W, Zhang W Y, Wang X, Wang M, Shen S W, Gao M F, Mei B, Wang H K, Tong J R, Cao L M, Feng D S, Sun B. Comparison of storage tolerance among 33 different genotypes of rice[J/OL]. Molecular Plant Breeding, 2024, https://link.cnki.net/urlid/46.1068.S.20240202.1646.010. (in Chinese with English abstract) | |
| [6] | Dong X Y, Fan S X, Liu J, Wang Q, Li M R, Jiang X, Liu Z Y, Yin Y C, Wang J Y. Identification of QTLs for seed storability in rice under natural aging conditions using two RILs with the same parent Shennong 265[J]. Journal of Integrative Agriculture, 2017, 16(5):1084-1092. |
| [7] | Jin J, Long W, Wang L, Liu X, Pan G, Xiang W, Li N, Li S. QTL mapping of seed vigor of backcross inbred lines derived from Oryza longistaminata under artificial aging[J]. Frontiers in Plant Science, 2018, 9: 1909. |
| [8] | Miura K, Lin Y, Yano M, Nagamine T. Mapping quantitative trait loci controlling seed longevity in rice (Oryza sativa L.)[J]. Theoretical and Applied Genetics, 2002, 104(6): 981-986. |
| [9] | Sasaki K, Takeuchi Y, Miura K, Yamaguchi T, Ando T, Ebitani T, Higashitani A, Yamaya T, Yano M, Sato T. Fine mapping of a major quantitative trait locus, qLG-9, that controls seed longevity in rice [J]. Theoretical and Applied Genetics, 2015, 128(4): 769-778. |
| [10] | Sasaki K, Fukuta K, Sato T. Mapping of quantitative trait loci controlling seed longevity of rice after various periods of seed storage[J]. Plant Breeding, 2005, 124(4): 361-366. |
| [11] | Xue Y, Zhang S Q, Yao Q H, Peng R H, Xiong A S, Li X, Zhu W M, Zhu Y Y, Zha D S. Identification of quantitative trait loci for seed storability in rice (Oryza sativa L.)[J]. Euphytica, 2008, 164(3): 739-744. |
| [12] | Jiang W, Lee J, Jin T M, Qiao Y, Piao R H, Jang S M, Woo M O, Kwon S W, Liu X H, Pan H Y, Du X, Kon H J. Identification of QTLs for seed germination capability after various storage periods using two RIL populations in rice[J]. Molecules and Cells, 2011, 31(4): 385-392. |
| [13] | Yuan Z, Fan K, Xia L, Ding X. Genetic dissection of seed storability and validation of candidate gene associated with antioxidant capability[J]. International Journal of Molecular Sciences, 2019, 20(18): 4442. |
| [14] | Yuan Z Y, Fan K, Wang Y T, Tian L, Zhang C P, Sun W Q, He H Z. OsGRETCHENHAGEN3-2 modulates rice seed storability via accumulation of abscisic acid[J]. Plant Physiology, 2021, 186(1): 469-482. |
| [15] | Zheng X H, Yuan Z Y, Yu Y Y, Yu S B, He H Z. OsCSD2 and OsCSD3 enhance seed storability by modulating antioxidant enzymes and abscisic acid in rice[J]. Plants, 2024, 13(2): 310. |
| [16] | 闫蕴韬, 何兮, 张海清, 贺记外. 水稻种子耐贮性研究进展[J]. 中国农学通报, 2022, 38(5): 1-8. |
| Yan Y T, He X, Zhang H Q, He J W. Advances in research on the storability of rice seeds[J]. Chinese Agricultural Science Bulletin, 2022, 38(5): 1-8. (in Chinese with English abstract) | |
| [17] | Xu H B, Wei Y D, Zhu Y S, Lian L, Xie H G, Cai Q H, Chen Q S, Lin Z P, Wang Z H, Xie H A, Zhang J F. Antisense suppression of LOX3 gene expression in rice endosperm enhances seed longevity[J]. Plant Biotechnology Journal, 2015, 13(4): 526-539. |
| [18] | Wang F X, Xu H B, Zhang L, Shi Y R, Song Y, Wang X Y, Cai Q H, He W, Xie H A, Zhang J F. The lipoxygenase OsLOX10 affects seed longevity and resistance to saline-alkaline stress during rice seedlings[J]. Plant Molecular Biology, 2023, 111(4): 415-428. |
| [19] | Wei Y, Xu H, Diao L, Zhu Y S, Xie H G, Cai Q H, Wu F X, Wang Z H, Zhang J F, Xie H A. Protein repair L-isoaspartyl methyltransferase 1 in rice improves seed longevity by preserving embryo vigor and viability[J]. Plant Molecular Biology, 2015, 89(5): 475-492. |
| [20] | Wang W Q, Xu D Y, Sui Y P, Ding X H, Song X J. A multiomic study uncovers a bZIP23-PER1A-mediated detoxification pathway to enhance seed vigor in rice[J]. Proceedings of the National Academy of Sciences of the United States of America, 2022, 119(9): e2026355119. |
| [21] | 江川, 李书柯, 李清华, 王金英. 水稻耐储藏性研究进展[J]. 江西农业学报, 2011, 23(10): 39-43. |
| Jiang C, Li S K, Li Q H, Wang J Y. Research progress in storability of rice[J]. Acta Agriculturae Jiangxi, 2011, 23(10): 39-43. (in Chinese with English abstract) | |
| [22] | Wang Y, Zhang Y J, Sun X X, He X T, Yang J L, Chen X F, Shi Z H, Song X L, Qiang S, Dai W M. Weedy rice de-domesticated from cultivated rice has evolved strong resistance to seed ageing[J]. Weed Research, 2021, 61(4): 396-405. |
| [23] | Zhou H, Li S, Liu J, Hu J, Le S, Li M. Identification and analysis of the genetic integrity of different types of rice resources through SSR markers[J]. Scientific Reports, 2023, 13:2428. |
| [24] | Ding G, Hu B, Zhou Y, Yang W, Zhao M, Xie J, Zhang F. Development and characterization of chromosome segment substitution lines derived from Oryza rufipogon in the background of the Oryza sativa indica restorer line R974[J]. Genes, 2022, 13(5): 735. |
| [25] | Zhou S Q, Huang K R, Zhou Y, Hu Y Q, Xiao Y C, Chen T, Yin M Q, Liu Y, Xu M L, Jiang X C. Degradome sequencing reveals an integrative miRNA-mediated gene interaction network regulating rice seed vigor[J]. BMC Plant Biology, 2022, 22: 269. |
| [26] | Meng L, Li H, Zhang L, Wang J. QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations[J]. The Crop Journal, 2015, 3: 269-283. |
| [27] | McCouch S R CGSNL. Gene nomenclature system for rice[J]. Rice, 2008, 1(1): 72-84. |
| [28] | Chen S F, Zhou Y Q, Chen Y R, Gu J. Fastp: an ultra-fast all-in-one FASTQ preprocessor[J]. Bioinformatics, 2018, 34(17): 884-890. |
| [29] | Jung Y, Han D. BWA-MEME: BWA-MEM emulated with a machine learning approach[J]. Bioinformatics. 2022, 38(9): 2404-2413. |
| [30] | McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo M A. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data[J]. Genome Research, 2010, 20(9): 1297-1303. |
| [31] | Cingolani P, Platts A, Wang L, Coon M, Nguyen T, Wang L, Land S J, Lu X, Ruden D M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila Melanogaster strain w1118; iso-2; iso-3[J]. Fly (Austin), 2012, 6(2): 80-92. |
| [32] | Hill J T, Demarest B L, Bisgrove B W, Gorsi B, Su Y C, Yost H J. MMAPPR: mutation mapping analysis pipeline for pooled RNA-seq[J]. Genome Research, 2013, 23(4): 687-697. |
| [33] | Zhao M M, Hu B L, Fan Y W, Ding G M, Yang W L, Chen Y, Chen Y H, Xie J K, Zhang F T. Identification, analysis, and confirmation of seed storability-related loci in Dongxiang wild rice (Oryza rufipogon Griff.)[J]. Genes, 2021, 12: 1831. |
| [34] | Choudhary P, Pramitha L, Aggarwal P R, Rana S, Vetriventhan M, Muthamilarasan M. Biotechnological interventions for improving the seed longevity in cereal crops: Progress and prospects[J]. Critical Reviews in Biotechnology, 2023, 43(2): 309-325. |
| [35] | 刘慧敏, 周杰强, 胡远艺, 田妍, 雷斌, 李建武, 魏中伟, 唐文帮. 水稻小粒不育系新组合卓两优1126的高产特征[J]. 中国水稻科学, 2024, 38(2): 160-171. |
| Liu H M, Zhou J Q, Hu Y Y, Tian Y, Lei B, Li J W, Wei Z W, Tang W B. Super-high yield characteristics of two-line hybrid rice Zhuoliangyou 1126[J]. Chinese Journal of Rice Science, 2024, 38(2): 160-171. (in Chinese with English abstract) | |
| [36] | Li C S, Shao G S, Wang L, Wang Z F, Mao Y J, Wang X Q, Zhang X H, Liu S T, Zhang H S. QTL identification and fine mapping for seed storability in rice (Oryza sativa L.)[J]. Euphytica, 2017, 213: 127. |
| [37] | Liu F Z, Li N N, Yu Y Y, Chen W, Yu S B, He H Z. Insights into the regulation of rice seed storability by seed tissue-specific transcriptomic and metabolic profiling[J]. Plants, 2022, 11(12): 1570. |
| [38] | Lin Q Y, Wang W Y, Ren Y K, Jiang Y M, Sun A L, Qian Y, Zhang Y F, He N Q, Hang N T, Liu Z, Li L F, Liu L L, Jiang L, Wan J M. Genetic dissection of seed storability using two different populations with a same parent rice cultivar N22[J]. Breeding Science, 2015, 65: 411-419. |
| [39] | Hang N T, Lin Q Y, Liu L L, Liu X, Liu S J, Wang W Y, Li L F, He N Q, Liu Z, Jiang L, Wan J M. Mapping QTLs related to rice seed storability under natural and artificial aging storage conditions[J]. Euphytica, 2015, 203: 673-681. |
| [40] | Wang W Q, Xu D Y, Sui Y P, Ding X H, Song X J. A multiomic study uncovers a bZIP23-PER1A-mediated detoxification pathway to enhance seed vigor in rice[J]. Proceedings of the National Academy of Sciences of the United States of America, 2022, 119(9): e2026355119. |
| [41] | Huang J X, Cai M H, Long Q Z, Liu L L, Lin Q Y, Jiang L, Chen S H, Wan J M. OsLOX2, a rice type I lipoxygenase, confers opposite effects on seed germination and longevity[J]. Transgenic Research, 2014, 23(4): 643-655. |
| [42] | 刘进, 姚晓云, 余丽琴, 李慧, 周慧颖, 王嘉宇, 黎毛毛. 水稻耐储藏特性三年动态鉴定与QTL分析[J]. 植物学报, 2019, 54(4): 464-473. |
| Liu J, Yao X Y, Yu L Q, Li H, Zhou H Y, Wang J Y, Li M M. Detection and analysis of dynamic quantitative trait loci at three years for seed storability in rice (Oryza sativa)[J]. Chinese Bulletin of Botany, 2019, 54(4): 464-473. (in Chinese with English abstract) | |
| [43] | Wang X, Ren P, Ji L, Zhu B, Xie G. OsVDE, a xanthophyll cycle key enzyme, mediates abscisic acid biosynthesis and negatively regulates salinity tolerance in rice[J]. Planta, 2021, 255: 6. |
| [44] | Ali F, Qanmber G, Li F, Wang Z. Updated role of ABA in seed maturation, dormancy, and germination[J]. Journal of Advanced Research, 2022, 35: 199-214. |
| [45] | Zheng J, Chen F Y, Wang Z, Cao H, Li X Y, Deng X, Soppe W J, Li Y, Liu Y X. A novel role for histone methyltransferase KYP⁄SUVH4 in the control of Arabidopsis primary seed dormancy[J]. New Phytologist, 2012, 193: 605-616. |
| [46] | Wang X, Zhou W, Lu Z H, Ouyang Y D, Yao J L. A lipid transfer protein, OsLTPL36, is essential for seed development and seed quality in rice[J]. Plant Science, 2015, 239: 200-208. |
| [47] | Li Y Y, Guo L N, Cui Y, Yan X, Ouyang J X, Li S B. Lipid transfer protein, OsLTPL18, is essential for grain weight and seed germination in rice[J]. Gene, 2023, 883:147671. |
| [48] | Wang Y, Hou Y, Qiu J, Wang H, Wang S, Tang L, Tong X, Zhang J. Abscisic acid promotes jasmonic acid biosynthesis via a SAPK10-bZIP72-AOC pathway to synergistically inhibit seed germination in rice (Oryza sativa)[J]. New Phytologist, 2020, 228: 1336-1353. |
| [49] | Zhou T S, Yu D, Wu L B, Xu Y S, Dun M J, Yuan D Y. Seed storability in rice: physiological bases, molecular mechanisms, and application to breeding[J]. Rice Science, 2024, 31(3): 1-16. |
| [50] | Chen B, Fu H, Gao J, Zhang Y, Huang W, Chen Z, Zhang Q, Yan S, Liu J. Identification of metabolomic biomarkers of seed vigor and aging in hybrid rice[J]. Rice, 2022, 15: 7. |
| [51] | Suzuki Y, Higo K, Hagiwara K, Ohtsubo K, Kobayashi A, Nozu Y. Production and use of monoclonal antibodies against rice embryo lipoxygenase-3[J]. Bioscience Biotechnology and Biochemistry, 1992, 56(4): 678-679. |
| [52] | Suzuki Y, Miura K, Shigemune A, Sasahara H, Ohta H, Uehara Y, Ishikawa T, Hamada S, Shirasawa K. Marker-assisted breeding of a LOX-3-null rice line with improved storability and resistance to preharvest sprouting[J]. Theoretical and Applied Genetics, 2015, 128(7): 1421-1430. |
| [53] | Lin Q Y, Jiang Y M, Sun A L, Cao P H, Li L F, Liu X, Tian Y L, HeJ, Liu S J. Fine mapping of qSS-9, a major and stable quantitative trait locus, for seed storability in rice[J]. Plant Breeding, 2015, 134(3): 293-299. |
| [1] | 杜彦修, 孙文玉, 袁泽科, 张倩倩, 李富豪, 李俊周, 孙红正. 利用QTL-Seq结合分子标记定位粳稻垩白粒率控制位点qChalk8[J]. 中国水稻科学, 2024, 38(6): 665-671. |
| [2] | 侯本福, 杨传铭, 张喜娟, 杨贤莉, 王立志, 王嘉宇, 李红宇, 姜树坤. 利用龙稻5号/中优早8号RIL群体定位粒形QTL[J]. 中国水稻科学, 2024, 38(1): 13-24. |
| [3] | 胡佳晓, 刘进, 崔迪, 勒思, 周慧颖, 韩冰, 孟冰欣, 余丽琴, 韩龙植, 马小定, 黎毛毛. 利用东乡野生稻染色体片段置换系鉴定穗部性状主效QTL[J]. 中国水稻科学, 2023, 37(6): 597-608. |
| [4] | 李杰, 田蓉蓉, 白天亮, 朱春艳, 宋佳伟, 田蕾, 马帅国, 吕建东, 胡慧, 王震宇, 罗成科, 张银霞, 李培富. 水稻回交群体剑叶性状综合评价及QTL定位[J]. 中国水稻科学, 2021, 35(6): 573-585. |
| [5] | 陈喜娜, 袁泽科, 胡珍珍, 赵全志, 孙红正. 利用QTL-Seq定位粳稻整精米率QTL[J]. 中国水稻科学, 2021, 35(5): 449-454. |
| [6] | 季芝娟, 曾宇翔, 梁燕, 杨长登. 水稻恶苗病抗性研究进展[J]. 中国水稻科学, 2021, 35(1): 1-10. |
| [7] | 吴婷, 李霞, 黄得润, 黄凤林, 肖宇龙, 胡标林. 应用东乡野生稻回交重组自交系分析水稻耐低氮产量相关性状QTL[J]. 中国水稻科学, 2020, 34(6): 499-511. |
| [8] | 张安鹏, 钱前, 高振宇. 水稻种子活力的研究进展[J]. 中国水稻科学, 2018, 32(3): 296-303. |
| [9] | 胡标林, 黄得润, 肖叶青, 何强生, 万勇, 樊叶杨. 应用东乡野生稻回交重组自交系群体分析糙米矿质含量QTL[J]. 中国水稻科学, 2018, 32(1): 43-50. |
| [10] | 曲丽君1,2,张宏军2,项超2,王辉3,夏加发3,李泽福3,高用明2,*,石英尧1,*. 杂交稻“大青棵”现象遗传基础剖析[J]. 中国水稻科学, 2013, 27(6): 559-568. |
| [11] | 江建华1,张晚霞1,刘晓丽1,刘强明1,卢超1,党小景1,赵其兵2,洪德林1,*. 多环境下粳稻株高动态QTL分析[J]. 中国水稻科学, 2012, 26(1): 55-64. |
| [12] | 徐建军1,2,赵强3,赵元凤1,朱磊1,徐辰武1,顾铭洪1,韩斌3,梁国华1,*. 利用重测序的水稻染色体片段代换系群体定位剑叶形态QTL[J]. 中国水稻科学, 2011, 25(5): 483-487. |
| [13] | 江建华,赵其兵,刘强明 ,陈 兰,陈甫龙,乔保健,洪德林,. 利用条件QTL定位发掘粳稻生育期和株高及单株有效穗数适用有利等位变异[J]. 中国水稻科学, 2011, 25(3): 277-283 . |
| [14] | 龚继平,吴方喜,吴跃进,郑家团,黄庭旭,王乌齐,张建福,谢华安,. 籼稻脂肪酶基因的遗传分析及定位[J]. 中国水稻科学, 2008, 22(2): 125-130 . |
| [15] | 杨空松,陈小荣,傅军如,朱昌兰,彭小松,贺晓鹏,贺浩华. 东乡野生稻育性恢复性的鉴定与遗传分析[J]. 中国水稻科学, 2007, 21(5): 487-492 . |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||