
Chinese Journal OF Rice Science ›› 2024, Vol. 38 ›› Issue (6): 627-637.DOI: 10.16819/j.1001-7216.2024.231210
• Research Papers • Previous Articles Next Articles
WANG Qing1,2, WANG Yanru2,3, ZHANG Xiuli1,2, LÜ Qiming1,2,*( )
)
Received:2023-12-10
Revised:2024-01-10
Online:2024-11-10
Published:2024-11-15
Contact:
*email: qmlv@hhrrc.ac.cn
汪晴1,2, 王艳茹2,3, 张秀丽1,2, 吕启明1,2,*( )
)
通讯作者:
*email: qmlv@hhrrc.ac.cn
基金资助:WANG Qing, WANG Yanru, ZHANG Xiuli, LÜ Qiming. Sequence Variation Analysis of the Parthenogeny-inducing Gene BBM1 in Rice[J]. Chinese Journal OF Rice Science, 2024, 38(6): 627-637.
汪晴, 王艳茹, 张秀丽, 吕启明. 水稻孤雌生殖诱导基因BBM1序列变异分析[J]. 中国水稻科学, 2024, 38(6): 627-637.
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.ricesci.cn/EN/10.16819/j.1001-7216.2024.231210
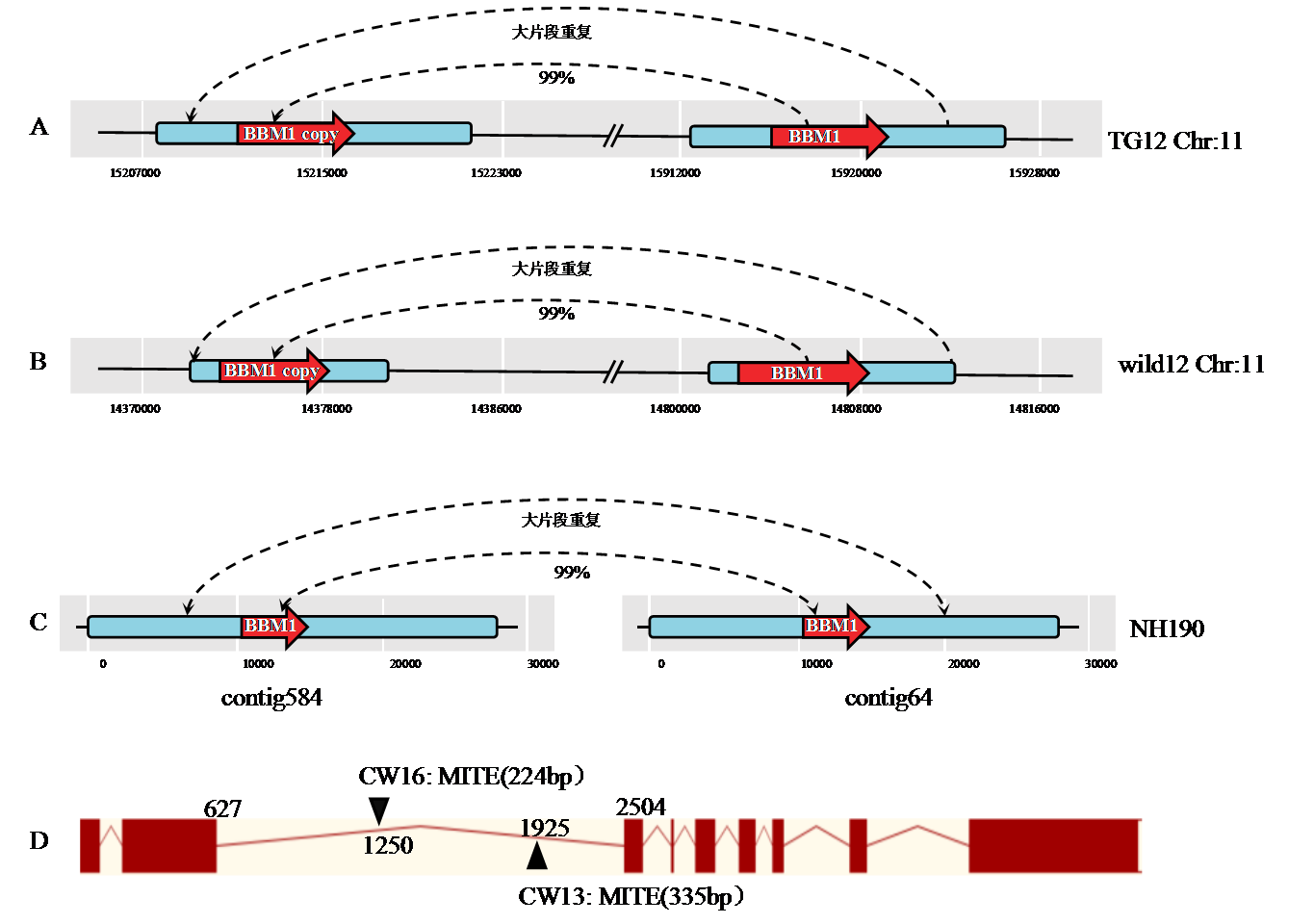
Fig. 1. Structural variation of BBM1 A, B, C show the BBM1 structural variants of TG12, wild12 and NH190, blue represents the BBM1 flanking large fragment repeat sequence and red represents the BBM1 gene sequence. D: Map of BBM1 gene structure of CW13 and CW16, burgundy represents exons, yellow represents introns, and ▲ represents MITE insertion sites.

Fig. 2. Nucleotide diversity of BBM1 sequences in different rice accessions A, B and C show the nucleotide polymorphisms of BBM1 sequence in wild rice, indica and japonica rice, respectively. The BBM1 exon is shown in burgundy and the BBM1 intron in orange.
| 水稻类型 Rice type | 对比序列数Number of sequences used | 多态性位点Number of polymorphic (segregating) sites | 突变总数 Total number of mutations | 单倍体数Number of haplotypes | 单倍型多样性Haplotype (gene) diversity | 单倍型多态性 方差 Variance of haplotype diversity | 单倍型多态性 标准差 Standard deviation of haplotype diversity | 核苷酸多态性Nucleotide diversity |
|---|---|---|---|---|---|---|---|---|
| 核心种质 Core collections | 383 | 268 | 285 | 69 | 0.868 | 0.0001 | 0.010 | 0.00506 |
| 野生稻 Wild rice | 30 | 196 | 200 | 29 | 0.998 | 0.00009 | 0.009 | 0.00623 |
| 籼稻 indica | 204 | 91 | 94 | 22 | 0.786 | 0.00045 | 0.021 | 0.00333 |
| 粳稻 japonica | 103 | 77 | 79 | 11 | 0.318 | 0.00359 | 0.060 | 0.00131 |
Table 1. Analysis of BBM1 sequence polymorphism in different rice accessions
| 水稻类型 Rice type | 对比序列数Number of sequences used | 多态性位点Number of polymorphic (segregating) sites | 突变总数 Total number of mutations | 单倍体数Number of haplotypes | 单倍型多样性Haplotype (gene) diversity | 单倍型多态性 方差 Variance of haplotype diversity | 单倍型多态性 标准差 Standard deviation of haplotype diversity | 核苷酸多态性Nucleotide diversity |
|---|---|---|---|---|---|---|---|---|
| 核心种质 Core collections | 383 | 268 | 285 | 69 | 0.868 | 0.0001 | 0.010 | 0.00506 |
| 野生稻 Wild rice | 30 | 196 | 200 | 29 | 0.998 | 0.00009 | 0.009 | 0.00623 |
| 籼稻 indica | 204 | 91 | 94 | 22 | 0.786 | 0.00045 | 0.021 | 0.00333 |
| 粳稻 japonica | 103 | 77 | 79 | 11 | 0.318 | 0.00359 | 0.060 | 0.00131 |

Fig. 5. Schematic representation of the BBM1 domain BBM1 contains two AP2 domains, the AP2 domain in red, the amino acid sequence in blue, and the amino acid position in numbers.
| 单倍型 Haplotype | 数量 Number | 16 | 67 | 69 | 107 | 124 | 142 | 146 | 147 | 147.1 | 258 | 351 | 445 | 493 | 521 | 537 | 561 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hap-1 | 109 | S | T | A | M | V | G | A | A | - | R | T | T | P | D | L | stop |
| Hap-36 | 92 | S | T | A | M | A | D | A | A | - | R | I | T | P | D | V | +26AA |
| Hap-37 | 51 | S | M | A | M | A | G | A | A | - | R | T | T | P | D | V | +26AA |
| Hap-22 | 29 | S | T | A | M | A | G | A | A | - | R | T | T | Q | D | V | +26AA |
| Hap-17 | 17 | S | T | A | M | A | G | A | A | - | R | T | T | P | D | V | +26AA |
| Hap-40 | 12 | S | T | A | M | A | D | A | A | - | R | I | I | P | D | V | +26AA |
| Hap-4 | 10 | S | T | A | M | A | G | - | - | - | R | T | T | P | N | V | +26AA |
| Hap-6 | 5 | S | T | A | M | A | G | - | - | - | R | T | T | P | D | V | +26AA |
| Hap-43 | 4 | Y | T | D | M | V | G | A | A | A | R | T | T | P | D | L | stop |
| Hap-23 | 3 | S | T | A | M | A | G | A | A | - | R | T | T | P | D | V | +26AA |
| Hap-8 | 2 | S | T | A | M | A | G | - | - | - | \ | \ | \ | \ | \ | \ | \ |
| Hap-44 | 2 | S | T | A | T | A | G | A | A | - | R | T | T | P | D | V | +26AA |
Table 2. Nonsynonymous mutation sites in different haplotypes
| 单倍型 Haplotype | 数量 Number | 16 | 67 | 69 | 107 | 124 | 142 | 146 | 147 | 147.1 | 258 | 351 | 445 | 493 | 521 | 537 | 561 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hap-1 | 109 | S | T | A | M | V | G | A | A | - | R | T | T | P | D | L | stop |
| Hap-36 | 92 | S | T | A | M | A | D | A | A | - | R | I | T | P | D | V | +26AA |
| Hap-37 | 51 | S | M | A | M | A | G | A | A | - | R | T | T | P | D | V | +26AA |
| Hap-22 | 29 | S | T | A | M | A | G | A | A | - | R | T | T | Q | D | V | +26AA |
| Hap-17 | 17 | S | T | A | M | A | G | A | A | - | R | T | T | P | D | V | +26AA |
| Hap-40 | 12 | S | T | A | M | A | D | A | A | - | R | I | I | P | D | V | +26AA |
| Hap-4 | 10 | S | T | A | M | A | G | - | - | - | R | T | T | P | N | V | +26AA |
| Hap-6 | 5 | S | T | A | M | A | G | - | - | - | R | T | T | P | D | V | +26AA |
| Hap-43 | 4 | Y | T | D | M | V | G | A | A | A | R | T | T | P | D | L | stop |
| Hap-23 | 3 | S | T | A | M | A | G | A | A | - | R | T | T | P | D | V | +26AA |
| Hap-8 | 2 | S | T | A | M | A | G | - | - | - | \ | \ | \ | \ | \ | \ | \ |
| Hap-44 | 2 | S | T | A | T | A | G | A | A | - | R | T | T | P | D | V | +26AA |
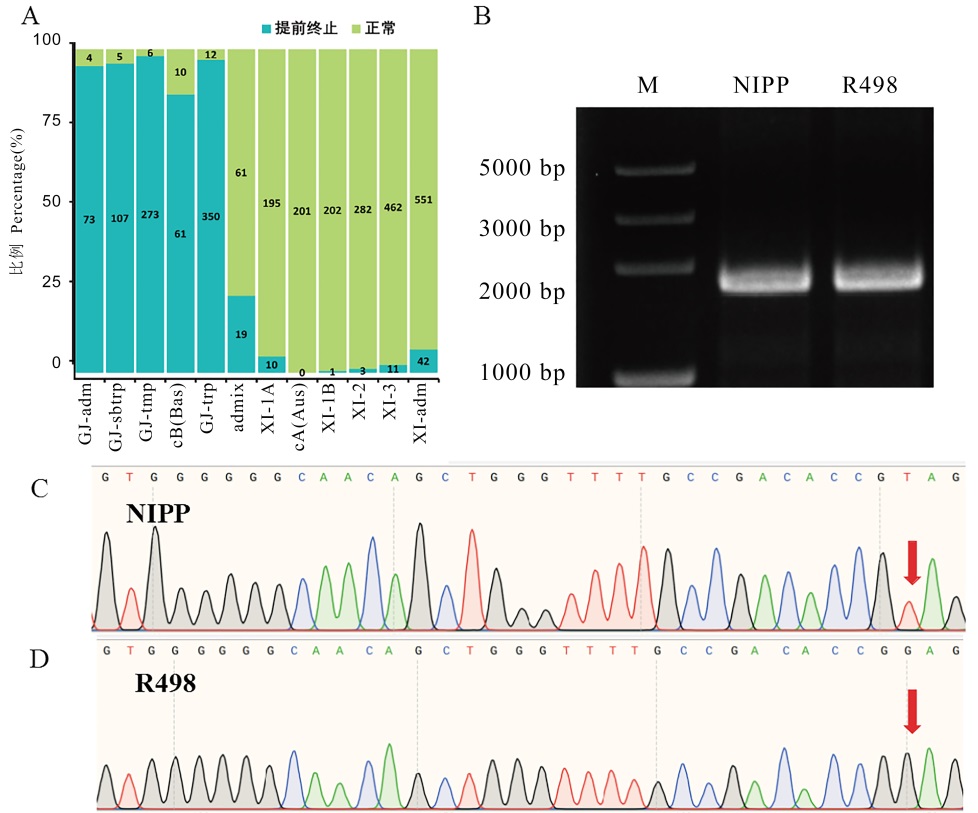
Fig. 6. Validation of different types of BBM1 sequences A, Distribution map of different BBM1 types in 2941 rice accessions, cyan indicates the occurrence of premature termination of BBM1, and green indicates normal BBM1; B, Results of agarose electrophoresis of full-length amplified BBM1 coding region sequences in NIPP and R498; C, Sequencing peak map of BBM1 coding region in NIPP; D, Sequencing peak map of BBM1 coding region in R498.
| [1] | Ye X X, Ma Y B, Sun B. Influence of soil type and genotype on Cd bioavailability and uptake by rice and implications for food safety[J]. Journal of Environmental Sciences, 2012, 24(9): 1647-1654. |
| [2] | 任光俊, 颜龙安, 谢华安. 三系杂交水稻育种研究的回顾与展望[J]. 科学通报, 2016, 61(35): 3748-3760. |
| Ren G J, Yan L A, Xie H A. Retrospective and perspective on indica three-line hybrid rice breeding research in China[J]. Chinese Science Bulletin, 2016, 61(35): 3748-3760. (in Chinese with English abstract) | |
| [3] | 袁隆平. 杂交水稻的育种战略设想[J]. 杂交水稻, 1987(1): 1-3. |
| Yuan L P. Strategy of hybrid rice breeding[J]. Hybrid Rice, 1987(1): 1-3. (in Chinese with English abstract) | |
| [4] | Bicknell R A, Koltunow A M. Understanding apomixis: Recent advances and remaining conundrums[J]. The Plant Cell, 2004, 16: 228-245. |
| [5] | Koltunow A M. Apomixis: Embryo sacs and embryos formed without meiosis or fertilization in ovules[J]. The Plant Cell, 1993, 5(10): 1425-1437. |
| [6] | 蔡得田, 陈冬玲. 论无融合生殖固定水稻杂种优势的策略[J]. 杂交水稻, 1989(6): 1-3. |
| Cai D T, Chen D L. Trying discussion in the tactics of fixing hybrid vigor of rice by apomixes[J]. Hybrid Rice, 1989(6): 1-3. (in Chinese with English abstract) | |
| [7] | 蔡得田, 马平福, 关和新, 姚家琳, 王灶安, 祝虹. 高频率无融合生殖水稻的研究[J]. 华中农业大学学报, 1991, 10(3): 223-227. |
| Cai D T, Ma P F, Gan H X, Yao J L, Wang Z A, Zhu H. The study of high frequency of apomictic rice material (HDAR). Journal of Huazhong Agricultural University, 1991, 10(3): 223-227. (in Chinese with English abstract) | |
| [8] | Jefferson R A. Apomixis: A social revolution for agriculture[J]. Biotechnology and Development Monitor, 1994, 19: 14-16. |
| [9] | Spillane C, Curtis M D, Grossniklaus U. Apomixis technology development-virgin births in farmers' fields?[J]. Nature Biotechnology, 2004, 22(6): 687-691. |
| [10] | Ondřej P, Michal S, Martin D. Reciprocal hybridization between diploid Ficaria calthifolia and tetraploid Ficaria verna subsp verna: Evidence from experimental crossing, genome size and molecular markers[J]. Botanical Journal of the Linnean Society, 2019, 189(3): 293-310. |
| [11] | Vijverberg K, Ozias-Akins P, Schranz M E. Identifying and engineering genes for parthenogenesis in plants[J]. Frontiers in Plant Science, 2019, 10: 128. |
| [12] | Brukhin V. Molecular and genetic regulation of apomixis[J]. Russian Journal of Genetics, 2017, 53(9): 943-964. |
| [13] | Mieulet D, Jolivet S, Rivard M, Cromer L, Vernet A, Mayonove P, Pereira L, Droc G, Courtois B, Guiderdoni E, Mercier R. Turning rice meiosis into mitosis[J]. Cell Research, 2016, 26(11): 1242-1254. |
| [14] | Rashid M, He G Y, Yang G X, Hussain J, Xu Y. AP2/ERF transcription factor in rice: Genome-wide canvas and syntenic relationships between monocots and eudicots[J]. Evolutionary Bioinformatics Online, 2012, 8: 321-355. |
| [15] | Xie W, Ding C, Hu H, Dong G, Zhang G, Qian Q, Ren D. Molecular events of rice AP2/ERF transcription factors[J]. International Journal of Molecular Sciences, 2022, 23(19): 12013. |
| [16] | Chen B, Maas L, Figueiredo D, Zhong Y, Reis R, Li M, Horstman A, Riksen T, Weemen M, Liu H, Siemons C, Chen S, Angenent G C, Boutilier K. BABY BOOM regulates early embryo and endosperm development[J]. Proceedings of the National Academy of Sciences of the United States of America, 2022, 119(25): e2201761119. |
| [17] | Anderson S N, Johnson C S, Chesnut J, Jones D S, Khanday I, Woodhouse M, Li C, Conrad L J, Russell S D, Sundaresan V. The zygotic transition is initiated in unicellular plant zygotes with asymmetric activation of parental genomes[J]. Developmental Cell, 2017, 43: 349-358. |
| [18] | Anderson S N, Johnson C S, Jones D S, Conrad L J, Gou X, Russell S D, Sundaresan V. Transcriptomes of isolated Oryza sativa gametes characterized by deep sequencing: Evidence for distinct sex-dependent chromatin and epigenetic states before fertilization[J]. Plant Journal, 2013, 76(5): 729-741. |
| [19] | Conner J A, Mookkan M, Huo H, Chae K, Ozias-Akins P. A parthenogenesis gene of apomict origin elicits embryo formation from unfertilized eggs in a sexual plant[J]. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112: 11205-11210. |
| [20] | Conner J A, Podio M, Ozias-Akins P. Haploid embryo production in rice and maize induced by PsASGR-BBML transgenes[J]. Plant Reproduction, 2017, 30: 41-52. |
| [21] | Khanday I, Skinner D, Yang B, Mercier R, Sundaresan V. A male-expressed rice embryogenic trigger redirected for asexual propagation through seeds[J]. Nature, 2019, 565(7737): 91-95. |
| [22] | Vernet A, Meynard D, Lian Q, Mieulet D, Gibert O, Bissah M, Rivallan R, Autran D, Leblanc O, Meunier A C, Frouin J, Taillebois J, Shankle K, Khanday I, Mercier R, Sundaresan V, Guiderdoni E. High-frequency synthetic apomixis in hybrid rice[J]. Nature Communications, 2022, 13(1): 7963. |
| [23] | Wei X, Liu C, Chen X, Lu H, Wang J, Yang S, Wang K. Synthetic apomixis with normal hybrid rice seed production[J]. Molecular Plant, 2023, 16(3): 489-492. |
| [24] | Wang C, Liu Q, Shen Y, Hua Y, Wang J, Lin J, Wu M, Sun T, Cheng Z, Mercier R, Wang K. Clonal seeds from hybrid rice by simultaneous genome engineering of meiosis and fertilization genes[J]. Nature Biotechnology, 2019, 37(3): 283-286. |
| [25] | Wang C C, Yu H, Huang J, Wang W S, Faruquee M, Zhang F, Zhao X Q, Fu B Y, Chen K, Zhang H L, Tai S S, Wei C, McNally K L, Alexandrov N, Gao X Y, Li J, Li Z K, Xu J L, Zheng T Q. Towards a deeper haplotype mining of complex traits in rice with RFGB v2.0[J]. Plant Biotechnology Journal, 2020, 18(1): 14-16. |
| [26] | Wang W, Mauleon R, Hu Z, Chebotarov D, Tai S, Wu Z, Li M, Zheng T, Fuentes R R, Zhang F, Mansueto L, Copetti D, Sanciangco M, Palis K C, Xu J, Sun C, Fu B, Zhang H, Gao Y, Zhao X, Shen F, Cui X, Yu H, Li Z, Chen M, Detras J, Zhou Y, Zhang X, Zhao Y, Kudrna D, Wang C, Li R, Jia B, Lu J, He X, Dong Z, Xu J, Li Y, Wang M, Shi J, Li J, Zhang D, Lee S, Hu W, Poliakov A, Dubchak I, Ulat V J, Borja F N, Mendoza J R, Ali J, Li J, Gao Q, Niu Y, Yue Z, Naredo M E B, Talag J, Wang X, Li J, Fang X, Yin Y, Glaszmann J C, Zhang J, Li J, Hamilton R S, Wing R A, Ruan J, Zhang G, Wei C, Alexandrov N, McNally K L, Li Z, Leung H. Genomic variation in 3,010 diverse accessions of Asian cultivated rice[J]. Nature, 2018, 557(7703): 43-49. |
| [27] | Lü Q, Li W, Sun Z, Ouyang N, Jing X, He Q, Wu J, Zheng J, Zheng J, Tang S, Zhu R, Tian Y, Duan M, Tan Y, Yu D, Sheng X, Sun X, Jia G, Gao H, Zeng Q, Li Y, Tang L, Xu Q, Zhao B, Huang Z, Lu H, Li N, Zhao J, Zhu L, Li D, Yuan L, Yuan D. Resequencing of 1,143 indica rice accessions reveals important genetic variations and different heterosis patterns[J]. Nature Communication, 2020, 11(1): 4778. |
| [28] | Yu H, Kou L, Li J. 10k-level integrated rice database shows power for exploiting rare variants[J]. Journal of Integrative Plant Biology, 2023, 65(12): 2539-2540. |
| [29] | Shang L, Li X, He H, Yuan Q, Song Y, Wei Z, Lin H, Hu M, Zhao F, Zhang C, Li Y, Gao H, Wang T, Liu X, Zhang H, Zhang Y, Cao S, Yu X, Zhang B, Zhang Y, Tan Y, Qin M, Ai C, Yang Y, Zhang B, Hu Z, Wang H, Lv Y, Wang Y, Ma J, Wang Q, Lu H, Wu Z, Liu S, Sun Z, Zhang H, Guo L, Li Z, Zhou Y, Li J, Zhu Z, Xiong G, Ruan J, Qian Q. A super pan-genomic landscape of rice[J]. Cell Research, 2022, 32(10): 878-896. |
| [30] | Qin P, Lu H, Du H, Wang H, Chen W, Chen Z, He Q, Ou S, Zhang H, Li X, Li X, Li Y, Liao Y, Gao Q, Tu B, Yuan H, Ma B, Wang Y, Qian Y, Fan S, Li W, Wang J, He M, Yin J, Li T, Jiang N, Chen X, Liang C, Li S. Pan-genome analysis of 33 genetically diverse rice accessions reveals hidden genomic variations[J]. Cell, 2021, 184(13): 3542-3558. |
| [31] | Zhang F, Xue H, Dong X, Li M, Zheng X, Li Z, Xu J, Wang W, Wei C. Long-read sequencing of 111 rice genomes reveals significantly larger pan-genomes[J]. Genome Research, 2022, 32(5): 853-863. |
| [32] | 项聪英, 蔡年俊, 李静, 羊健, 陈剑平, 张恒木. 一个水稻小热休克蛋白基因的克隆和鉴定[J]. 中国水稻科学, 2016, 30(6): 587-592. |
| Xiang C Y, Cai N J, Li J, Yang J, Chen J P, Zhang H M. Cloning and characterization of a small heat shock protein (SHSP) gene in rice plant[J]. Chinese Journal of Rice Science, 2016, 30(6): 587-592. (in Chinese with English abstract) | |
| [33] | Tamura K, Stecher G, Kumar S. MEGA11: Molecular evolutionary genetics analysis version 11[J]. Molecular Biology and Evolution, 2021, 38(7): 3022-3027. |
| [34] | Rozas J, Ferrer-Mata A, Sánchez-DelBarrio J C, Guirao-Rico S, Librado P, Ramos-Onsins S E, Sánchez-Gracia A. DnaSP 6: DNA sequence polymorphism analysis of large data sets[J]. Molecular Biology Evolution, 2017, 34(12): 3299-3302. |
| [35] | Kawahara Y, de la Bastide M, Hamilton J P, Kanamori H, McCombie W R, Ouyang S, Schwartz D C, Tanaka T, Wu J, Zhou S, Childs K L, Davidson R M, Lin H, Quesada-Ocampo L, Vaillancourt B, Sakai H, Lee S S, Kim J, Numa H, Itoh T, Buell C R, Matsumoto T. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data[J]. Rice, 2013, 6(1): 4. |
| [36] | Chen C, Chen H, Zhang Y, Thomas H R, Frank M H, He Y, Xia R. TBtools: An integrative toolkit developed for interactive analyses of big biological data[J]. Molecular Plant, 2020, 13(8): 1194-1202. |
| [37] | 赵凤利. 水稻全基因组拷贝数变异研究揭示复制基因的非对称进化[D]. 北京: 中国农业科学院, 2021. |
| Zhao F L. A genome-wide survey of copy number variations reveals an asymmetric evolution of duplicated genes in rice[D]. Beijing: Agricultural Genomics Institute Graduate School, 2021. (in Chinese with English abstract) | |
| [38] | Guillet-Claude C, Birolleau-Touchard C, Manicacci D, Rogowsky P M, Rigau J, Murigneux A, Martinant J P, Barrière Y. Nucleotide diversity of the ZmPox3 maize peroxidase gene: Relationships between a MITE insertion in exon 2 and variation in forage maize digestibility[J]. BMC Genetics, 2004, 16(5): 19. |
| [1] |
SUI Jingjing, ZHAO Guilong, JIN Xin, BU Qingyun, TANG Jiaqi.
Advances in Molecular and Physiological Mechanisms of Cold Tolerance Regulation of Rice at the Booting Stage [J]. Chinese Journal OF Rice Science, 2025, 39(1): 1-10. |
| [2] |
REN Ningning, SUN Yongjian, SHEN Congcong, ZHU Shuangbing, LI Huiju, ZHANG Zhiyuan, CHEN Kai.
Research Progress in Rice Mesocotyl [J]. Chinese Journal OF Rice Science, 2025, 39(1): 11-23. |
| [3] |
XIAO Wuwei, ZHU Chenguang, WANG Fei, XIONG Dongliang, HUANG Jianliang, PENG Shaobing, CUI Kehui.
Research Progress in Rice Quality of Ratoon Rice [J]. Chinese Journal OF Rice Science, 2025, 39(1): 33-46. |
| [4] |
CHEN Zhihui, TAO Yajun, FAN Fangjun, XU Yang, WANG Fangquan, LI Wenqi, GULINAER·Bahetibieke, JIANG Yanjie, ZHU Jianping, LI Xia, YANG Jie.
Development and Application of a Functional Marker for Heading Date Gene Hd6 in Rice [J]. Chinese Journal OF Rice Science, 2025, 39(1): 47-54. |
| [5] |
HU Fengyue, WANG Jian, WANG Chun, WANG Kejian, LIU Chaolei.
Generation of Rice DMP1, DMP2 and DMP3 Mutants and Identification of Their Haploid Induction Ability [J]. Chinese Journal OF Rice Science, 2025, 39(1): 55-66. |
| [6] |
YANG Chuanming, WANG Lizhi, ZHANG Xijuan, YANG Xianli, WANG Yangyang, HOU Benfu, CUI Shize2, 4, LI Qingchao, LIU Kai4, MA Rui, FENG Yanjiang, LAI Yongcai, LI Hongyu, JIANG Shukun.
Analysis of QTL Controlling Cold Tolerance at Seedling Stage by Using a High-Density SNP Linkage Map in japonica Rice [J]. Chinese Journal OF Rice Science, 2025, 39(1): 82-91. |
| [7] |
CHEN Shurong, ZHU Lianfeng, QIN Birong, WANG Jie, Zhu Xuhua, TIAN Wenhao, ZHU Chunquan, CAO Xiaochuang, KONG Yali, ZHANG Junhua, JIN Qianyu.
Effects of Nitrification Inhibitors on Rice Growth, Yield and Nitrogen Use Efficiency Under Oxygenated Irrigation [J]. Chinese Journal OF Rice Science, 2025, 39(1): 92-100. |
| [8] |
WU Meng, NI Chuan, KANG Yuying, MAO Yuxin, YE Miao, ZHANG Zujian.
Inter-varietal Differences in Early Tillering Characteristics and Their Responses to Nitrogen [J]. Chinese Journal OF Rice Science, 2025, 39(1): 101-114. |
| [9] |
WANG Xiaoxi, CAI Chuang, SONG Lian, ZHOU Wei, YANG Xiong, GU Xinyue ZHU Chunwu.
Effect of Free-air CO2 Enrichment and Temperature Increase on Grain Quality of Rice Cultivar Yangdao 6 [J]. Chinese Journal OF Rice Science, 2025, 39(1): 115-127. |
| [10] |
JIANG Min, WANG Guanglun, LI Minglu, MIAO Bo, LI Mingxuan, SHI Chunlin.
Risk Assessment and Dynamic Early Warming of Heat Damage in Rice Based on Simulation Model [J]. Chinese Journal OF Rice Science, 2025, 39(1): 128-142. |
| [11] | YANG Jie, YANG Changdeng, ZENG Yuxiang, HOU Yuxuan, CHEN Tianxiao, LIANG Yan. Research Progress in Mining and Utilization of Rice Blast Resistance Genes [J]. Chinese Journal OF Rice Science, 2024, 38(6): 591-603. |
| [12] | FENG Xiangqian, WANG Aidong, HONG Weiyuan, LI Ziqiu, QIN Jinhua, ZHAN Lichuan, CHEN Lipeng, ZHANG Yunbo, WANG Danying, CHEN Song. Research Progress in Rice Yield Estimation Method Based on Low-altitude Unmanned Aerial Vehicle Remote Sensing [J]. Chinese Journal OF Rice Science, 2024, 38(6): 604-616. |
| [13] | YE Miao, MAO Yuxin, ZHANG Dehai, KANG Yuying, YUAN Rong, ZHANG Zujian. Advances in Leaf and Canopy Eco-physiological Characteristics of High Photosynthetic Efficiency Rice Varieties and Their Regulation Mechanisms by Nitrogen [J]. Chinese Journal OF Rice Science, 2024, 38(6): 617-626. |
| [14] | ZHONG Zhihu, QIN Lu, LI Zhili, YANG Zhen, HE Xiaopeng, CAI Yicong. Genome-wide Identification and Comprehensive Analysis of IDD Gene Family in Rice [J]. Chinese Journal OF Rice Science, 2024, 38(6): 638-652. |
| [15] | LI Wei, XU Xia, BIAN Ying, ZHANG Xiaobo, FAN Jiongjiong, CHENG Benyi, YANG Shihua, WU Jianli, WEI Xin, ZENG Bo, GONG Junyi. Cytoplasmic Source Analysis of Sterile Lines from 5460 Three-line Hybrid Rice Varieties [J]. Chinese Journal OF Rice Science, 2024, 38(6): 653-664. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||