
Chinese Journal OF Rice Science ›› 2024, Vol. 38 ›› Issue (6): 638-652.DOI: 10.16819/j.1001-7216.2024.240101
• Research Papers • Previous Articles Next Articles
ZHONG Zhihu, QIN Lu, LI Zhili, YANG Zhen, HE Xiaopeng, CAI Yicong*( )
)
Received:2024-01-01
Revised:2024-03-18
Online:2024-11-10
Published:2024-11-15
Contact:
*email: caiyicongcyc@163.com
通讯作者:
*email: caiyicongcyc@163.com
基金资助:ZHONG Zhihu, QIN Lu, LI Zhili, YANG Zhen, HE Xiaopeng, CAI Yicong. Genome-wide Identification and Comprehensive Analysis of IDD Gene Family in Rice[J]. Chinese Journal OF Rice Science, 2024, 38(6): 638-652.
钟智慧, 秦璐, 黎志力, 杨珍, 贺晓鹏, 蔡怡聪. 水稻IDD基因家族的全基因组鉴定及综合分析[J]. 中国水稻科学, 2024, 38(6): 638-652.
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.ricesci.cn/EN/10.16819/j.1001-7216.2024.240101
| 引物名称 Primer name | 正向引物序列 Forward sequence (5’-3’) | 反向引物序列 Revers sequence(5’-3’) |
|---|---|---|
| Actin-RT | CGGGAAATTGTGAGGGACAT | AGGAAGGCTGGAAGAGGACC |
| OsID1-RT | CTCTTCTCCAGGAAGGACAGCC | GTAGTAGTGATGCTGCTGCTGTTG |
| OsIDD1-RT | GTTCTGGTCGCGCTGGAAC | GACACAATCATTAGGAGCGGCA |
| OsIDD2-RT | CGGCAACCCAGATCCTGAG | CTGGTCTCGCTGGAACCC |
| OsIDD3-RT | GGAAACCCAAATCCAGATGCG | CCGCTGGAACCCCTTGTTG |
| OsIDD4-RT | CCTTACCAGACCCGGACGC | GTTCTGCTCCCGCTGGAAC |
| OsIDD5-RT | GGATGAGCAAGAGGTGGTTG | ATCCAGCAGGTTTGAGTCCT |
| OsIDD6-RT | CCCGGGAATCCAAATCCTGATGC | CTGCTCCCTCTGGAACCCC |
| OsIDD7-RT | GGAAACCCAGATCCAGATGTTGAG | GGAGGTTCTGGTCTCTCTGGAAG |
| OsIDD8-RT | CGGCACACCAGATCCGG | CAGGTTCTGGTCTCGCTGG |
| OsIDD9-RT | GTCTTCTCCAGGCGCGAC | CAGAGCGGAGGTGACCG |
| OsIDD10-RT | CAACCAACCCAAACCCGGAC | GGTTCTGCTCCCGCTGG |
| OsIDD11-RT | GCAACCCAGACCCGGAAG | CTGCAGGTTCTGGTCCCTC |
| OsIDD12-RT | CAGATCACCTGCTACAGCTG | TGACGACGGACGAGACGTA |
| OsIDD13-RT | GCACGCCAGACCCGGAC | CTGCAGGTTCTGGTCTCGC |
| OsIDD14-RT | GCGTCTTCTCCCGAGTGG | GGATGACCGGCAGCGTC |
Table 1. Primers used in the study.
| 引物名称 Primer name | 正向引物序列 Forward sequence (5’-3’) | 反向引物序列 Revers sequence(5’-3’) |
|---|---|---|
| Actin-RT | CGGGAAATTGTGAGGGACAT | AGGAAGGCTGGAAGAGGACC |
| OsID1-RT | CTCTTCTCCAGGAAGGACAGCC | GTAGTAGTGATGCTGCTGCTGTTG |
| OsIDD1-RT | GTTCTGGTCGCGCTGGAAC | GACACAATCATTAGGAGCGGCA |
| OsIDD2-RT | CGGCAACCCAGATCCTGAG | CTGGTCTCGCTGGAACCC |
| OsIDD3-RT | GGAAACCCAAATCCAGATGCG | CCGCTGGAACCCCTTGTTG |
| OsIDD4-RT | CCTTACCAGACCCGGACGC | GTTCTGCTCCCGCTGGAAC |
| OsIDD5-RT | GGATGAGCAAGAGGTGGTTG | ATCCAGCAGGTTTGAGTCCT |
| OsIDD6-RT | CCCGGGAATCCAAATCCTGATGC | CTGCTCCCTCTGGAACCCC |
| OsIDD7-RT | GGAAACCCAGATCCAGATGTTGAG | GGAGGTTCTGGTCTCTCTGGAAG |
| OsIDD8-RT | CGGCACACCAGATCCGG | CAGGTTCTGGTCTCGCTGG |
| OsIDD9-RT | GTCTTCTCCAGGCGCGAC | CAGAGCGGAGGTGACCG |
| OsIDD10-RT | CAACCAACCCAAACCCGGAC | GGTTCTGCTCCCGCTGG |
| OsIDD11-RT | GCAACCCAGACCCGGAAG | CTGCAGGTTCTGGTCCCTC |
| OsIDD12-RT | CAGATCACCTGCTACAGCTG | TGACGACGGACGAGACGTA |
| OsIDD13-RT | GCACGCCAGACCCGGAC | CTGCAGGTTCTGGTCTCGC |
| OsIDD14-RT | GCGTCTTCTCCCGAGTGG | GGATGACCGGCAGCGTC |
| 名称 Name | 染色体 Chr. | 基因长度 Gene length | 氨基酸数目 No. of amino acids | 分子量 Molecular weight | 理论等电点 Theoretical pI | 蛋白质不稳定 指数 Instability index | 脂溶指数 Aliphatic index | 亲水性平均系数 Grand average of hydropathicity | 亚细胞定位预测 Predicted subcellular localization |
|---|---|---|---|---|---|---|---|---|---|
| OsID1 | 10 | 1230 | 409 | 44267.44 | 8.04870033 | 51.19 | 51.19 | −0.467 | 细胞核Nucleus |
| OsIDD1 | 3 | 1659 | 552 | 57540.01 | 8.55090046 | 75.58 | 75.58 | −0.486 | 细胞核Nucleus |
| OsIDD2 | 1 | 1464 | 487 | 51815.90 | 8.60980034 | 48.96 | 48.96 | −0.527 | 细胞核Nucleus |
| OsIDD3 | 9 | 1608 | 535 | 54893.05 | 9.41469955 | 46.65 | 46.65 | −0.460 | 细胞核Nucleus |
| OsIDD4 | 2 | 1848 | 615 | 63597.71 | 8.63129997 | 46.29 | 46.29 | −0.497 | 细胞核Nucleus |
| OsIDD5 | 7 | 1902 | 633 | 67636.09 | 6.01849985 | 55.08 | 55.08 | −0.567 | 细胞核Nucleus |
| OsIDD6 | 8 | 1659 | 552 | 57262.74 | 9.38479996 | 49.81 | 49.81 | −0.500 | 细胞核Nucleus |
| OsIDD7 | 2 | 1479 | 492 | 54491.89 | 7.31449986 | 59.23 | 59.23 | −0.697 | 细胞核Nucleus |
| OsIDD8 | 1 | 1488 | 495 | 51394.20 | 8.89400005 | 49.83 | 49.83 | −0.293 | 细胞核Nucleus |
| OsIDD9 | 1 | 1431 | 476 | 50107.14 | 7.95529985 | 52.62 | 52.62 | −0.265 | 细胞核Nucleus |
| OsIDD10 | 4 | 1842 | 613 | 63814.37 | 9.93620014 | 52.40 | 52.4 | −0.476 | 细胞核Nucleus |
| OsIDD11 | 1 | 1614 | 537 | 57410.90 | 7.97130013 | 62.67 | 62.67 | −0.703 | 细胞核Nucleus |
| OsIDD12 | 8 | 1602 | 533 | 56426.28 | 9.51509953 | 62.47 | 62.47 | −0.467 | 细胞核Nucleus |
| OsIDD13 | 9 | 1515 | 504 | 53574.19 | 8.50090027 | 62.67 | 62.67 | −0.459 | 细胞核Nucleus |
| OsIDD14 | 3 | 1317 | 438 | 45959.66 | 10.12440010 | 70.46 | 70.46 | −0.509 | 细胞核Nucleus |
Table 2. Physical and chemical properties of OsIDD
| 名称 Name | 染色体 Chr. | 基因长度 Gene length | 氨基酸数目 No. of amino acids | 分子量 Molecular weight | 理论等电点 Theoretical pI | 蛋白质不稳定 指数 Instability index | 脂溶指数 Aliphatic index | 亲水性平均系数 Grand average of hydropathicity | 亚细胞定位预测 Predicted subcellular localization |
|---|---|---|---|---|---|---|---|---|---|
| OsID1 | 10 | 1230 | 409 | 44267.44 | 8.04870033 | 51.19 | 51.19 | −0.467 | 细胞核Nucleus |
| OsIDD1 | 3 | 1659 | 552 | 57540.01 | 8.55090046 | 75.58 | 75.58 | −0.486 | 细胞核Nucleus |
| OsIDD2 | 1 | 1464 | 487 | 51815.90 | 8.60980034 | 48.96 | 48.96 | −0.527 | 细胞核Nucleus |
| OsIDD3 | 9 | 1608 | 535 | 54893.05 | 9.41469955 | 46.65 | 46.65 | −0.460 | 细胞核Nucleus |
| OsIDD4 | 2 | 1848 | 615 | 63597.71 | 8.63129997 | 46.29 | 46.29 | −0.497 | 细胞核Nucleus |
| OsIDD5 | 7 | 1902 | 633 | 67636.09 | 6.01849985 | 55.08 | 55.08 | −0.567 | 细胞核Nucleus |
| OsIDD6 | 8 | 1659 | 552 | 57262.74 | 9.38479996 | 49.81 | 49.81 | −0.500 | 细胞核Nucleus |
| OsIDD7 | 2 | 1479 | 492 | 54491.89 | 7.31449986 | 59.23 | 59.23 | −0.697 | 细胞核Nucleus |
| OsIDD8 | 1 | 1488 | 495 | 51394.20 | 8.89400005 | 49.83 | 49.83 | −0.293 | 细胞核Nucleus |
| OsIDD9 | 1 | 1431 | 476 | 50107.14 | 7.95529985 | 52.62 | 52.62 | −0.265 | 细胞核Nucleus |
| OsIDD10 | 4 | 1842 | 613 | 63814.37 | 9.93620014 | 52.40 | 52.4 | −0.476 | 细胞核Nucleus |
| OsIDD11 | 1 | 1614 | 537 | 57410.90 | 7.97130013 | 62.67 | 62.67 | −0.703 | 细胞核Nucleus |
| OsIDD12 | 8 | 1602 | 533 | 56426.28 | 9.51509953 | 62.47 | 62.47 | −0.467 | 细胞核Nucleus |
| OsIDD13 | 9 | 1515 | 504 | 53574.19 | 8.50090027 | 62.67 | 62.67 | −0.459 | 细胞核Nucleus |
| OsIDD14 | 3 | 1317 | 438 | 45959.66 | 10.12440010 | 70.46 | 70.46 | −0.509 | 细胞核Nucleus |
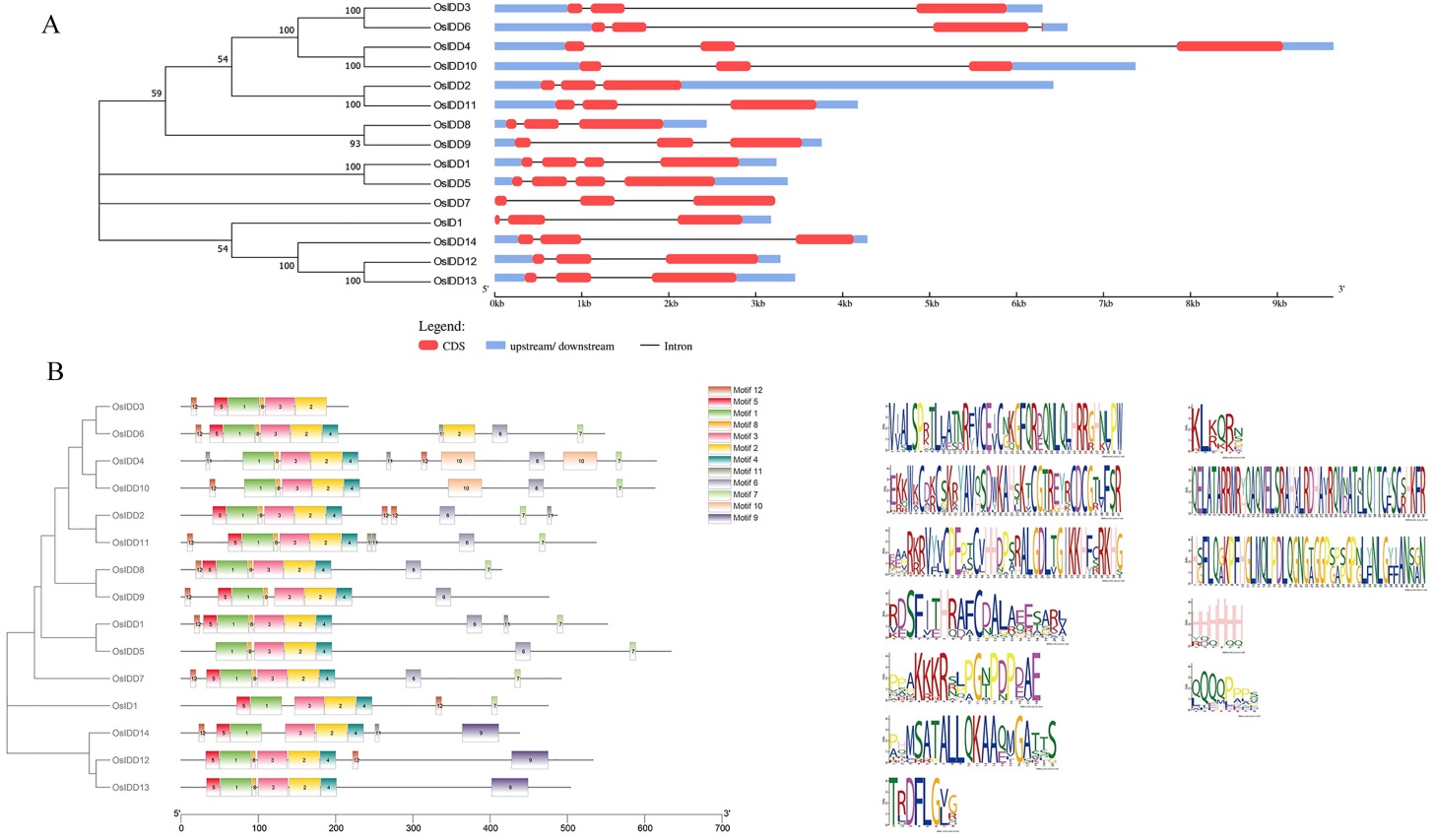
Fig. 2. Schematic graph of OsIDD gene structure and OsIDD Motif analysis A, Analysis of gene structure for OsIDD family; B, Analysis of OsIDD-protein motif domain.

Fig. 3. Sequence alignment of OsIDD proteins The blue boxes indicate nuclear localization signal region (NLS); the red boxes indicate zinc finger domains (C, H residues).
| 基因名称 Gene name | α-螺旋 α-helix | β-折叠 β-bridge | 延伸链 Extended strand | β-转角 β-turn | 无规则卷曲 Random coil |
|---|---|---|---|---|---|
| OsID1 | 92 (22.49%) | 0 | 89 (21.76%) | 33 (8.07%) | 195 (47.68%) |
| OsIDD1 | 78 (14.13%) | 0 | 66 (11.96%) | 31 (5.62%) | 377 (68.30%) |
| OsIDD2 | 149 (30.60%) | 0 | 42 (8.62%) | 32 (6.57%) | 264 (54.21%) |
| OsIDD3 | 97 (18.13%) | 0 | 71 (13.27%) | 30 (5.61%) | 337 (62.99%) |
| OsIDD4 | 182 (29.59%) | 0 | 77 (12.52%) | 48 (7.80%) | 308 (50.08%) |
| OsIDD5 | 132 (20.85%) | 0 | 97 (15.32%) | 50 (7.90%) | 354 (55.92%) |
| OsIDD6 | 150 (27.17%) | 0 | 87 (15.76%) | 48 (8.70%) | 267 (48.37%) |
| OsIDD7 | 99 (20.12%) | 0 | 48 (9.76%) | 16 (3.25%) | 329 (66.87%) |
| OsIDD8 | 147 (29.70%) | 0 | 64 (12.93%) | 42 (8.48%) | 242 (48.89%) |
| OsIDD9 | 112 (23.53%) | 0 | 68 (14.29%) | 45 (9.45%) | 251 (52.73%) |
| OsIDD10 | 172 (28.06%) | 0 | 82 (13.38%) | 37 (6.04%) | 322 (52.53%) |
| OsIDD11 | 195 (36.31%) | 0 | 58 (10.80%) | 46 (8.57%) | 238 (44.32%) |
| OsIDD12 | 200 (37.52%) | 0 | 69 (12.95%) | 33 (6.19%) | 231 (43.34%) |
| OsIDD13 | 225 (44.64%) | 0 | 40 (7.94%) | 13 (2.58%) | 226 (44.84%) |
| OsIDD14 | 190 (43.38%) | 0 | 41 (9.36%) | 35 (7.99%) | 172 (39.27%) |
Table 3. Secondary structure prediction analysis of the OsIDD proteins
| 基因名称 Gene name | α-螺旋 α-helix | β-折叠 β-bridge | 延伸链 Extended strand | β-转角 β-turn | 无规则卷曲 Random coil |
|---|---|---|---|---|---|
| OsID1 | 92 (22.49%) | 0 | 89 (21.76%) | 33 (8.07%) | 195 (47.68%) |
| OsIDD1 | 78 (14.13%) | 0 | 66 (11.96%) | 31 (5.62%) | 377 (68.30%) |
| OsIDD2 | 149 (30.60%) | 0 | 42 (8.62%) | 32 (6.57%) | 264 (54.21%) |
| OsIDD3 | 97 (18.13%) | 0 | 71 (13.27%) | 30 (5.61%) | 337 (62.99%) |
| OsIDD4 | 182 (29.59%) | 0 | 77 (12.52%) | 48 (7.80%) | 308 (50.08%) |
| OsIDD5 | 132 (20.85%) | 0 | 97 (15.32%) | 50 (7.90%) | 354 (55.92%) |
| OsIDD6 | 150 (27.17%) | 0 | 87 (15.76%) | 48 (8.70%) | 267 (48.37%) |
| OsIDD7 | 99 (20.12%) | 0 | 48 (9.76%) | 16 (3.25%) | 329 (66.87%) |
| OsIDD8 | 147 (29.70%) | 0 | 64 (12.93%) | 42 (8.48%) | 242 (48.89%) |
| OsIDD9 | 112 (23.53%) | 0 | 68 (14.29%) | 45 (9.45%) | 251 (52.73%) |
| OsIDD10 | 172 (28.06%) | 0 | 82 (13.38%) | 37 (6.04%) | 322 (52.53%) |
| OsIDD11 | 195 (36.31%) | 0 | 58 (10.80%) | 46 (8.57%) | 238 (44.32%) |
| OsIDD12 | 200 (37.52%) | 0 | 69 (12.95%) | 33 (6.19%) | 231 (43.34%) |
| OsIDD13 | 225 (44.64%) | 0 | 40 (7.94%) | 13 (2.58%) | 226 (44.84%) |
| OsIDD14 | 190 (43.38%) | 0 | 41 (9.36%) | 35 (7.99%) | 172 (39.27%) |
| 基因名称Gene symbol | 登录号 Accession number | 基因名称 Gene symbol | 登录号 Accession number | 基因名称 Gene symbol | 登录号 Accession number | 基因名称 Gene symbol | 登录号 Accession number |
|---|---|---|---|---|---|---|---|
| OsID1 | LOC_Os10g28330 | AtIDD1 | At5G66730 | AtIDD16 | AT1G25250 | ZmIDD14 | GRMZM2G141031 |
| OsIDD1 | LOC_Os03g10140 | AtIDD2 | At3G50700 | ZmID1 | GRMZM2G011357 | ZmIDD15 | GRMZM2G123094 |
| OsIDD2 | LOC_Os01g09850 | AtIDD3 | At1G03840 | ZmIDD1 | GRMZM2G171073 | ZmIDD16 | GRMZM2G074032 |
| OsIDD3 | LOC_Os09g38340 | AtIDD4 | AT2G02080 | ZmIDD2 | GRMZM2G143723 | ZmIDD17 | GRMZM2G110107 |
| OsIDD4 | LOC_Os02g45054 | AtIDD5 | AT2G02070 | ZmIDD3 | GRMZM5G828179 | ZmIDD18 | GRMZM2G465595 |
| OsIDD5 | LOC_Os07g39310 | AtIDD6 | AT1G14580 | ZmIDD4 | GRMZM2G151309 | ZmIDDp1 | GRMZM2G179677 |
| OsIDD6 | LOC_Os08g44050 | AtIDD7 | At1G55110 | ZmIDD5 | GRMZM2G046290 | ZmIDDveg7 | GRMZM2G042666 |
| OsIDD7 | LOC_Os02g31890 | AtIDD8 | At5G44160 | ZmIDD6 | GRMZM2G035625 | ZmIDDveg9 | GRMZM2G129261 |
| OsIDD8 | LOC_Os01g14010 | AtIDD9 | At3G45260 | ZmIDD7 | GRMZM2G320287 | ZmIDDp10 | GRMZM2G090595 |
| OsIDD9 | LOC_Os01g70870 | AtIDD10 | At5G03150 | ZmIDD8 | GRMZM2G022213 | ||
| OsIDD10 | LOC_Os04g47860 | AtIDD11 | At3G13810 | ZmIDD9 | GRMZM5G884137 | ||
| OsIDD11 | LOC_Os01g39110 | AtIDD12 | At4G02670 | ZmIDD10 | GRMZM2G058197 | ||
| OsIDD12 | LOC_Os08g36390 | AtIDD13 | At5G60470 | ZmIDD11 | GRMZM2G177693 | ||
| OsIDD13 | LOC_Os09g27650 | AtIDD14 | AT1G68130 | ZmIDD12 | GRMZM2G027333 | ||
| OsIDD14 | LOC_Os03g13400 | AtIDD15 | AT2G01940 | ZmIDD13 | GRMZM2G021587 |
Table 4. Summary of the IDD gene members in Oryza sativa, Arabidopsis, and Zea mays
| 基因名称Gene symbol | 登录号 Accession number | 基因名称 Gene symbol | 登录号 Accession number | 基因名称 Gene symbol | 登录号 Accession number | 基因名称 Gene symbol | 登录号 Accession number |
|---|---|---|---|---|---|---|---|
| OsID1 | LOC_Os10g28330 | AtIDD1 | At5G66730 | AtIDD16 | AT1G25250 | ZmIDD14 | GRMZM2G141031 |
| OsIDD1 | LOC_Os03g10140 | AtIDD2 | At3G50700 | ZmID1 | GRMZM2G011357 | ZmIDD15 | GRMZM2G123094 |
| OsIDD2 | LOC_Os01g09850 | AtIDD3 | At1G03840 | ZmIDD1 | GRMZM2G171073 | ZmIDD16 | GRMZM2G074032 |
| OsIDD3 | LOC_Os09g38340 | AtIDD4 | AT2G02080 | ZmIDD2 | GRMZM2G143723 | ZmIDD17 | GRMZM2G110107 |
| OsIDD4 | LOC_Os02g45054 | AtIDD5 | AT2G02070 | ZmIDD3 | GRMZM5G828179 | ZmIDD18 | GRMZM2G465595 |
| OsIDD5 | LOC_Os07g39310 | AtIDD6 | AT1G14580 | ZmIDD4 | GRMZM2G151309 | ZmIDDp1 | GRMZM2G179677 |
| OsIDD6 | LOC_Os08g44050 | AtIDD7 | At1G55110 | ZmIDD5 | GRMZM2G046290 | ZmIDDveg7 | GRMZM2G042666 |
| OsIDD7 | LOC_Os02g31890 | AtIDD8 | At5G44160 | ZmIDD6 | GRMZM2G035625 | ZmIDDveg9 | GRMZM2G129261 |
| OsIDD8 | LOC_Os01g14010 | AtIDD9 | At3G45260 | ZmIDD7 | GRMZM2G320287 | ZmIDDp10 | GRMZM2G090595 |
| OsIDD9 | LOC_Os01g70870 | AtIDD10 | At5G03150 | ZmIDD8 | GRMZM2G022213 | ||
| OsIDD10 | LOC_Os04g47860 | AtIDD11 | At3G13810 | ZmIDD9 | GRMZM5G884137 | ||
| OsIDD11 | LOC_Os01g39110 | AtIDD12 | At4G02670 | ZmIDD10 | GRMZM2G058197 | ||
| OsIDD12 | LOC_Os08g36390 | AtIDD13 | At5G60470 | ZmIDD11 | GRMZM2G177693 | ||
| OsIDD13 | LOC_Os09g27650 | AtIDD14 | AT1G68130 | ZmIDD12 | GRMZM2G027333 | ||
| OsIDD14 | LOC_Os03g13400 | AtIDD15 | AT2G01940 | ZmIDD13 | GRMZM2G021587 |
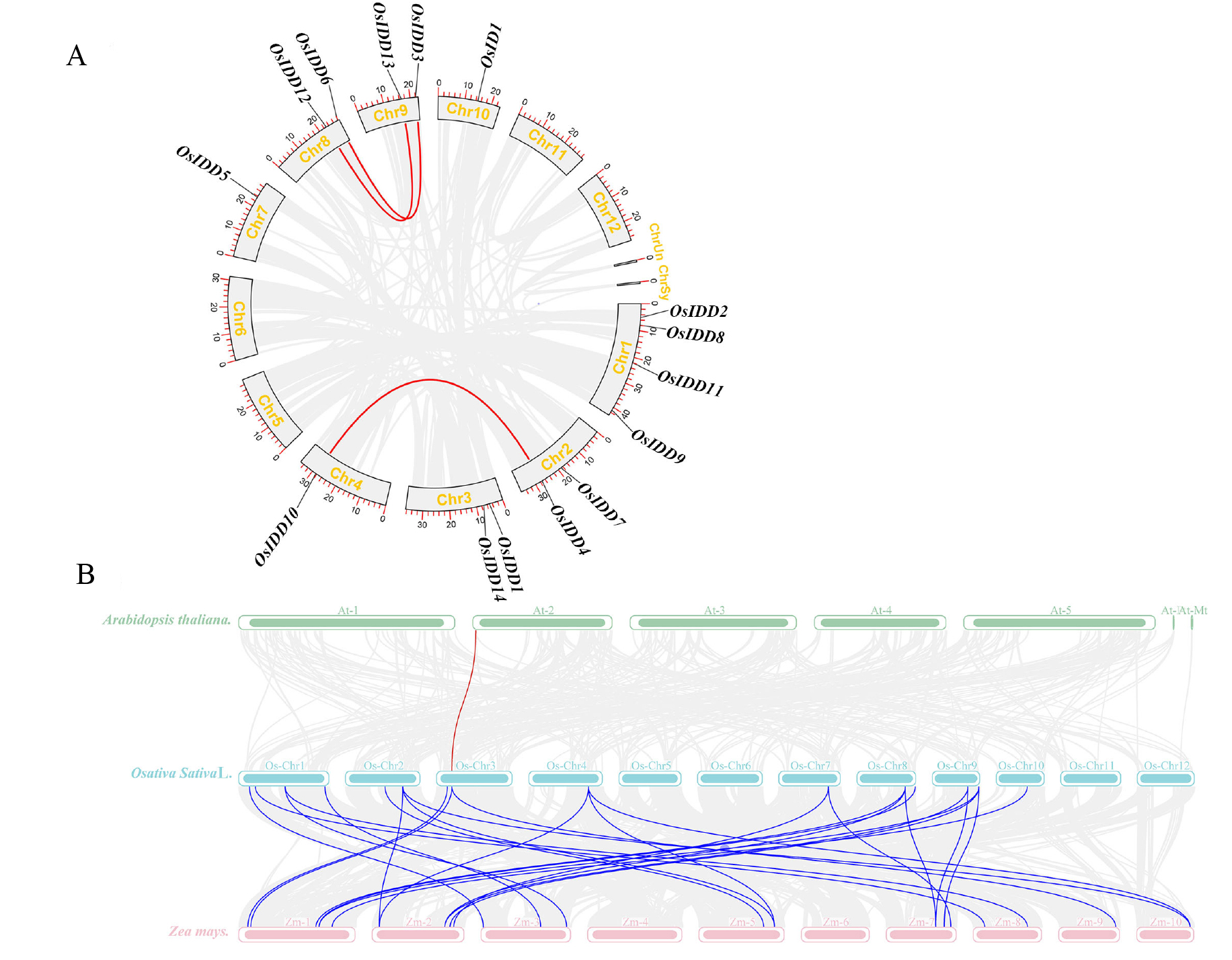
Fig. 6. Segmental gene pairs of OsIDD genes and collinearity analysis of IDD gene with Arabidopsis and Zea mays A, Segmental gene pairs of OsIDDs genes; B, Collinearity analysis of OsIDDs and AtIDDs, ZmIDDs genes; Lines represents segmental gene pairs.
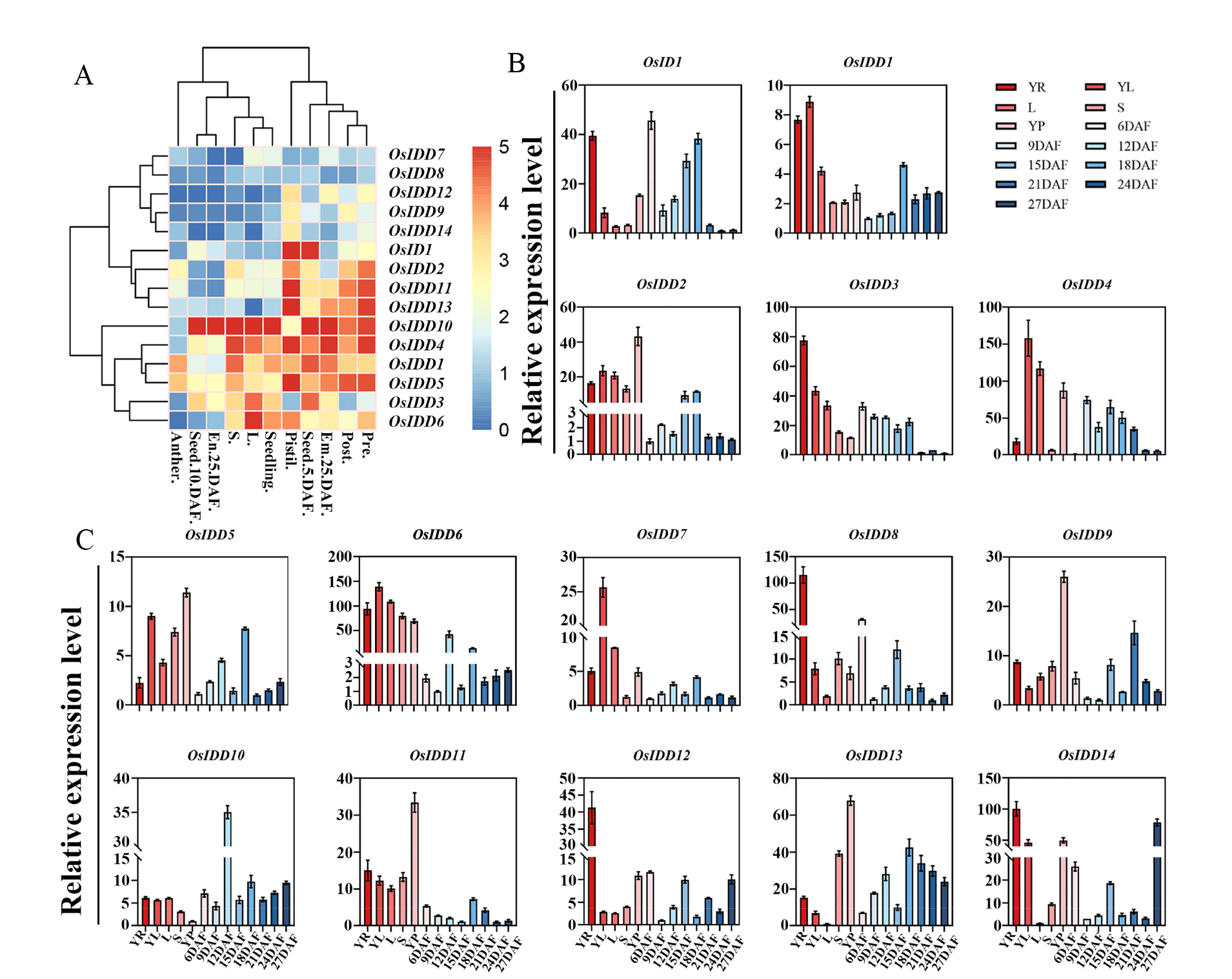
Fig. 7. Expression pattern of OsIDD A, OsIDDs expression level thermogram; B~C, OsIDDs expression level in rice tissues; YR, Root at seedling stage with three fully expanded leaves; YL; Leaves at seedling stage with three fully expanded leaves; L, Leaf at booting stage; S, Stem at booting stage; YP, Young spikelet; 6DAF, Developed endosperm for 6 days; 9DAF, Developed endosperm for 9 days; 12DAF, Developed endosperm for 12 days; 15DAF, Developed endosperm for 15 days; 18DAF, Developed endosperm for 18 days; 21DAF, Developed endosperm for 21 days; 24DAF, Developed endosperm for 24 days; 27DAF, Developed endosperm for 27 days. Data are mean ± SD from three biological replicates.
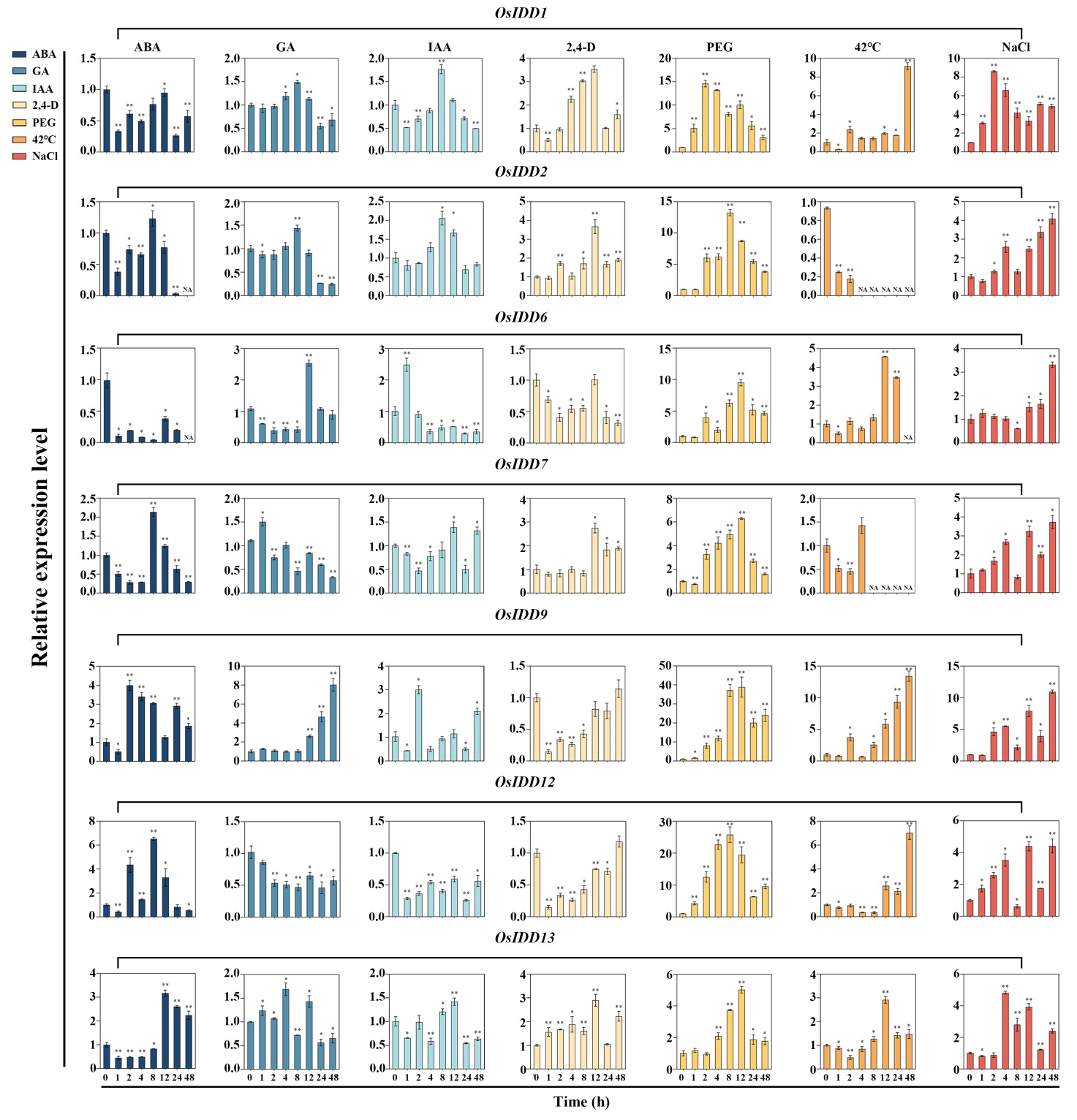
Fig. 9. Expression analysis of OsIDD genes (part) under different hormone treatments and different stress conditions ABA, GA, IAA, 2, 4-D were different hormones treatments; PEG, 42℃, and NaCl were different stress conditions. Mean ± SD (n=3); The asterisks on the top of the columns indicate significant differences from the value at 0 h (*, P < 0.05; **, P < 0.01).
| [1] | 孙燕, 苟德明, 李文鑫. C2H2型锌指蛋白研究进展[J]. 生命的化学, 2001 (6): 473-475. |
| Sun Y, Gou D M, Li W X. Advances in the study of C2H2 type zinc finger proteins[J]. Chemistry of Life, 2001(6): 473-475. (in Chinese with English abstract) | |
| [2] | Macpherson S, Larochelle M, Turcotte B. A fungal family of transcriptional regulators: the zinc cluster proteins[J]. Microbiology and Molecular Biology Reviews, 2006, 70(3): 583-604. |
| [3] | Lee M S, Gippert G P, Soman K V, Case D A, Wright P E. Three-dimensional solution structure of a single zinc finger DNA-binding domain[J]. Science, 1989, 245(4918): 635-637. |
| [4] | Párraga G, Horvath S J, Eisen A, Taylor W E, Hood L, Young E T, Klevit R E. Zinc-dependent structure of a single-finger domain of yeast ADR1[J]. Science, 1988, 241(4872): 1489-1492. |
| [5] | Xu Q K, Yu H P, Xia S S, Cui Y J, Yu X Q, Liu H, Zeng D L, Hu J, Zhang Q, Gao Z Y, Zhang G H, Zhu L, Shen L, Guo L B, Rao Y C, Qian Q, Ren D Y. The C2H2 zinc-finger protein LACKING RUDIMENTARY GLUME 1 regulates spikelet development in rice[J]. Science Bulletin, 2020, 65(9): 753-764. |
| [6] | 侯思宇, 孙朝霞, 郭彬, 王玉国, 李贵全, 韩渊怀. 大豆两个C2H2型转录因子基因序列特征及表达分析[J]. 植物生理学报, 2014, 50(5): 665-674. |
| Hou S Y, Sun Z X, Guo B, Wang Y G, Li G Q, Han Y H. Cloning and expression analysis of two C2H2 transcription factors in soybean[J]. Plant Physiology Journal, 2014, 50(5): 665-674. (in Chinese with English abstract) | |
| [7] | 孟繁君, 陈明, 徐长营, 朴秀吉, 汪可心, 葛善欣, 金玄吉. 玉米C2H2型锌指蛋白基因ZFP225的鉴定、生物信息学分析与克隆[J]. 作物杂志, 2014 (1): 49-53. |
| Meng F J, Chen M, Xu C Y, Piao X J, Wang K X, Ge S X, Jin X J. Identification, bioinformatic analysis and cloning of type C2H2 zinc finger protein gene ZFP225 in maize[J]. Crops, 2014(1): 49-53. (in Chinese with English abstract) | |
| [8] | Sakamoto H, Araki T, Meshi T, Iwabuchi M. Expression of a subset of the Arabidopsis Cys(2)/His(2)-type zinc-finger protein gene family under water stress[J]. Gene, 2000, 248(1-2): 23-32. |
| [9] | Park S J, Kim S L, Lee S, Je B I, Piao H L, Park S H, Kim C M, Ryu C H, Park S H, Xuan Y H, Colasanti J, An G, Han C D. Rice Indeterminate 1 (OsId1) is necessary for the expression of Ehd1 (Early heading date 1) regardless of photoperiod[J]. The Plant Journal, 2008, 56(6): 1018-1029. |
| [10] | Matsubara K, Yamanouchi U, Wang Z X, Minobe Y, Izawa T, Yano M. Ehd2, a rice ortholog of the maize INDETERMINATE1 gene, promotes flowering by up-regulating Ehd1[J]. Plant Physiology, 2008, 148(3): 1425-1435. |
| [11] | Colasanti J, Tremblay R, Wong A Y, Coneva V, Kozaki A, Mable B K. The maize INDETERMINATE1 flowering time regulator defines a highly conserved zinc finger protein family in higher plants[J]. BMC Genomics, 2006, 7: 158. |
| [12] | Colasanti J, Yuan Z, Sundaresan V. The indeterminate gene encodes a zinc finger protein and regulates a leaf-generated signal required for the transition to flowering in maize[J]. Cell, 1998, 93(4): 593-603. |
| [13] | Wu C Y, You C J, Li C S, Long T, Chen G X, Byrne M E, Zhang Q F. RID1, encoding a Cys2/His2-type zinc finger transcription factor, acts as a master switch from vegetative to floral development in rice[J]. Proceedings of the National Academy of Sciences of the United States of America, 2008, 105(35): 12915-12920. |
| [14] | Coneva V, Guevara D, Rothstein S J, Colasanti J. Transcript and metabolite signature of maize source leaves suggests a link between transitory starch to sucrose balance and the autonomous floral transition[J]. Journal of Experimental Botany, 2012, 63(2): 5079-5092. |
| [15] | Sakamoto H, Maruyama K, Sakuma Y, Meshi T, Iwabuchi M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought, cold, and high-salinity stress conditions[J]. Plant Physiology, 2004, 136(1): 2734-2746. |
| [16] | Gontarek B C, Neelakandan A K, Wu H, Becraft P W. NKD transcription factors are central regulators of maize endosperm development[J]. The Plant Cell, 2016, 28(12): 2916-2936. |
| [17] | Coelho C P, Huang P, Lee D Y, Brutnell T P. Making roots, shoots, and seeds: IDD gene family diversification in plants[J]. Trends in Plant Science, 2018, 23(1): 66-78. |
| [18] | Morita M T, Sakaguchi K, Kiyose S I, Taira K, Kato T, Nakamura M, Tasaka M. A C2H2‐type zinc finger protein, SGR5, is involved in early events of gravitropism in Arabidopsis inflorescence stems[J]. The Plant Journal, 2006, 47(4): 619-628. |
| [19] | Cui D Y, Zhao J B, Jing Y J, Fan M Z, Liu J, Wang Z C, Xin W, Hu Y X. The Arabidopsis IDD14, IDD15, and IDD16 cooperatively regulate lateral organ morphogenesis and gravitropism by promoting auxin biosynthesis and transport[J]. PLoS Genetics, 2013, 9(9): e1003759. |
| [20] | Feurtado J A, Huang D, Wicki-Stordeur L, Hemstock L E, Potentier M S, Tsang E W, Cutler A J. The Arabidopsis C2H2 zinc finger INDETERMINATE DOMAIN1/ ENHYDROUS promotes the transition to germination by regulating light and hormonal signaling during seed maturation[J]. The Plant Cell, 2011, 23(5): 1772-1794. |
| [21] | Patterson K, Cakmak T, Cooper A, Lager I, Rasmusson A G, Escobar M A. Distinct signalling pathways and transcriptome response signatures differentiate ammonium- and nitrate-supplied plants[J]. Plant, Cell & Environment, 2010, 33(9): 1486-1501. |
| [22] | Seo P J, Ryu J, Kang S K, Park C M. Modulation of sugar metabolism by an INDETERMINATE DOMAIN transcription factor contributes to photoperiodic flowering in Arabidopsis[J]. The Plant Journal, 2011, 65(3): 418-429. |
| [23] | Matsubara K, Yamanouchi U, Wang Z X, Minobe Y, Izawa T, Yano M. Ehd2, a rice ortholog of the maize INDETERMINATE1 gene, promotes flowering by up-regulating Ehd1[J]. Plant Physiology, 2008, 148(3): 1425-1435. |
| [24] | Deng L, Li L M, Zhang S, Shen J Q, Li S B, Hu S F, Peng Q, Xiao J H, Wu C Y. Suppressor of rid1 (SID1) shares common targets with RID1 on florigen genes to initiate floral transition in rice[J]. PLOS Genetics, 2017, 13(2): e1006642. |
| [25] | Dou M Z, Cheng S, Zhao B T, Xuan Y H, Shao M L. The Indeterminate Domain Protein ROC1 regulates chilling tolerance via activation of DREB1B/CBF1 in rice[J]. International Journal of Molecular Sciences, 2016, 17(3): 233. |
| [26] | Liu J M, Park S J, Huang J, Lee E J, Xuan Y H, Je B I, Kumar V, Priatama R A, Raj K V, Kim S H, Min M K, Cho J H, Kim T H, Chandran A K, Jung K H, Takatsuto S, Fujioka S, Han C D. Loose Plant Architecture1 (LPA1) determines lamina joint bending by suppressing auxin signalling that interacts with C-22-hydroxylated and 6-deoxo brassinosteroids in rice[J]. Journal of Experimental Botany, 2016, 67(6): 1883-1895. |
| [27] | Wu X R, Tang D, Li M, Wang K J, Cheng Z K. Loose Plant Architecture1, an INDETERMINATE DOMAIN Protein involved in shoot gravitropism, regulates plant architecture in rice[J]. Plant Physiology, 2013, 161(1): 317-329. |
| [28] | Cui Z B, Xue C Y, Mei Q, Xuan Y H. Malectin Domain Protein Kinase (MDPK) promotes rice resistance to sheath blight via IDD12, IDD13, and IDD14[J]. International Journal of Molecular Sciences, 2022, 23(15): 8214. |
| [29] | Livak K J, Schmittgen T D. Analysis of relative gene expression data using realtime quantitative pcr and the 2-△△CT method[J]. Methods, 2001, 25(4): 402-408. |
| [30] | Malheiros R S P, Costa L C, Ávila R T, Pimenta T M, Teixeira L S, Brito F A L, Zsögön A, Araújo W L, Ribeiro D M. Selenium downregulates auxin and ethylene biosynthesis in rice seedlings to modify primary metabolism and root architecture[J]. Planta, 2019, 250(1): 333-345. |
| [31] | Fu Z Z, Yu J, Cheng X W, Zong X, Xu J, Chen M J, Li Z Y, Zhang D B, Liang W Q. The rice Basic Helix-Loop- Helix Transcription Factor TDR INTERACTING PROTEIN2 is a central switch in early anther development[J]. The Plant Cell, 2014, 26(4): 1512-1524. |
| [32] | Ko S S, Li M J, Sun-Ben Ku M, Ho Y C, Lin Y J, Chuang M H, Hsing H X, Lien Y C, Yang H T, Chang H C, Chan M T. The bHLH142 transcription factor coordinates with TDR1 to modulate the expression of EAT1 and regulate pollen development in rice[J]. The Plant Cell, 2014, 26(6): 2486-2504. |
| [33] | 杜逸凡, 方希林, 曾红丽, 于玉凤, 张露倩, 王悦. 外源多效唑, ABA, 2, 4-D和NAA对水稻秧苗生长特性的影响[J]. 分子植物育种, 2020, 18(8): 2687-2694. |
| Du Y F, Fang X L, Zeng H L, Yu Y F, Zhang L Q, Wang Y. Effects of exogenous Paclobutrazol, ABA, 2, 4-D, and NAA on growth characteristics of rice seedlings[J]. Molecular Plant Breeding, 18(8): 2687-2694. (in Chinese with English abstract) | |
| [34] | Huang S Y, Liu M M, Chen G L, Si F F, Fan F F, Guo Y, Yuan L, Yang F, Li S Q. Favorable QTLs from Oryza longistaminata improve rice drought resistance[J]. BMC Plant Biology, 2022, 22(1): 136. |
| [35] | Song Y, Zhang C J, Ge W N, Zhang Y F, Burlingame A L, Guo Y. Identification of NaCl stress-responsive apoplastic proteins in rice shoot stems by 2D-DIGE[J]. Journal of Proteomics, 2011, 74(7): 1045-1067. |
| [36] | Kozaki A, Hake S, Colasanti J. The maize ID1 flowering time regulator is a zinc finger protein with novel DNA binding properties[J]. Nucleic Acids Research, 2004, 32(5): 1710-1720. |
| [37] | Seo P J, Kim M J, Ryu J Y, Jeong E Y, Park C M. Two splice variants of the IDD14 transcription factor competitively form nonfunctional heterodimers which may regulate starch metabolism[J]. Nature Communications, 2011, 2: 303. |
| [38] | Sun Q, Li T Y, Li D D, Wang Z Y, Li S, Li D P, Han X, Liu J M, Xuan Y H. Overexpression of Loose Plant Architecture 1 increases planting density and resistance to sheath blight disease via activation of PIN‐FORMED 1a in rice[J]. Plant Biotechnology Journal, 2019, 17(5): 855-857. |
| [39] | Lu Y Z, Feng Z, Meng Y L, Bian L Y, Xie H, Mysore K S, Liang J S. SLENDER RICE1 and Oryza sativa INDETERMINATE DOMAIN 2 regulating OsmiR396 are involved in stem elongation[J]. Plant Physiology, 2020, 182(4): 2213-2227. |
| [1] |
SUI Jingjing, ZHAO Guilong, JIN Xin, BU Qingyun, TANG Jiaqi.
Advances in Molecular and Physiological Mechanisms of Cold Tolerance Regulation of Rice at the Booting Stage [J]. Chinese Journal OF Rice Science, 2025, 39(1): 1-10. |
| [2] |
REN Ningning, SUN Yongjian, SHEN Congcong, ZHU Shuangbing, LI Huiju, ZHANG Zhiyuan, CHEN Kai.
Research Progress in Rice Mesocotyl [J]. Chinese Journal OF Rice Science, 2025, 39(1): 11-23. |
| [3] |
XIAO Wuwei, ZHU Chenguang, WANG Fei, XIONG Dongliang, HUANG Jianliang, PENG Shaobing, CUI Kehui.
Research Progress in Rice Quality of Ratoon Rice [J]. Chinese Journal OF Rice Science, 2025, 39(1): 33-46. |
| [4] |
CHEN Zhihui, TAO Yajun, FAN Fangjun, XU Yang, WANG Fangquan, LI Wenqi, GULINAER·Bahetibieke, JIANG Yanjie, ZHU Jianping, LI Xia, YANG Jie.
Development and Application of a Functional Marker for Heading Date Gene Hd6 in Rice [J]. Chinese Journal OF Rice Science, 2025, 39(1): 47-54. |
| [5] |
HU Fengyue, WANG Jian, WANG Chun, WANG Kejian, LIU Chaolei.
Generation of Rice DMP1, DMP2 and DMP3 Mutants and Identification of Their Haploid Induction Ability [J]. Chinese Journal OF Rice Science, 2025, 39(1): 55-66. |
| [6] |
YANG Chuanming, WANG Lizhi, ZHANG Xijuan, YANG Xianli, WANG Yangyang, HOU Benfu, CUI Shize2, 4, LI Qingchao, LIU Kai4, MA Rui, FENG Yanjiang, LAI Yongcai, LI Hongyu, JIANG Shukun.
Analysis of QTL Controlling Cold Tolerance at Seedling Stage by Using a High-Density SNP Linkage Map in japonica Rice [J]. Chinese Journal OF Rice Science, 2025, 39(1): 82-91. |
| [7] |
CHEN Shurong, ZHU Lianfeng, QIN Birong, WANG Jie, Zhu Xuhua, TIAN Wenhao, ZHU Chunquan, CAO Xiaochuang, KONG Yali, ZHANG Junhua, JIN Qianyu.
Effects of Nitrification Inhibitors on Rice Growth, Yield and Nitrogen Use Efficiency Under Oxygenated Irrigation [J]. Chinese Journal OF Rice Science, 2025, 39(1): 92-100. |
| [8] |
WU Meng, NI Chuan, KANG Yuying, MAO Yuxin, YE Miao, ZHANG Zujian.
Inter-varietal Differences in Early Tillering Characteristics and Their Responses to Nitrogen [J]. Chinese Journal OF Rice Science, 2025, 39(1): 101-114. |
| [9] |
WANG Xiaoxi, CAI Chuang, SONG Lian, ZHOU Wei, YANG Xiong, GU Xinyue ZHU Chunwu.
Effect of Free-air CO2 Enrichment and Temperature Increase on Grain Quality of Rice Cultivar Yangdao 6 [J]. Chinese Journal OF Rice Science, 2025, 39(1): 115-127. |
| [10] |
JIANG Min, WANG Guanglun, LI Minglu, MIAO Bo, LI Mingxuan, SHI Chunlin.
Risk Assessment and Dynamic Early Warming of Heat Damage in Rice Based on Simulation Model [J]. Chinese Journal OF Rice Science, 2025, 39(1): 128-142. |
| [11] | YANG Jie, YANG Changdeng, ZENG Yuxiang, HOU Yuxuan, CHEN Tianxiao, LIANG Yan. Research Progress in Mining and Utilization of Rice Blast Resistance Genes [J]. Chinese Journal OF Rice Science, 2024, 38(6): 591-603. |
| [12] | FENG Xiangqian, WANG Aidong, HONG Weiyuan, LI Ziqiu, QIN Jinhua, ZHAN Lichuan, CHEN Lipeng, ZHANG Yunbo, WANG Danying, CHEN Song. Research Progress in Rice Yield Estimation Method Based on Low-altitude Unmanned Aerial Vehicle Remote Sensing [J]. Chinese Journal OF Rice Science, 2024, 38(6): 604-616. |
| [13] | YE Miao, MAO Yuxin, ZHANG Dehai, KANG Yuying, YUAN Rong, ZHANG Zujian. Advances in Leaf and Canopy Eco-physiological Characteristics of High Photosynthetic Efficiency Rice Varieties and Their Regulation Mechanisms by Nitrogen [J]. Chinese Journal OF Rice Science, 2024, 38(6): 617-626. |
| [14] | WANG Qing, WANG Yanru, ZHANG Xiuli, LÜ Qiming. Sequence Variation Analysis of the Parthenogeny-inducing Gene BBM1 in Rice [J]. Chinese Journal OF Rice Science, 2024, 38(6): 627-637. |
| [15] | LI Wei, XU Xia, BIAN Ying, ZHANG Xiaobo, FAN Jiongjiong, CHENG Benyi, YANG Shihua, WU Jianli, WEI Xin, ZENG Bo, GONG Junyi. Cytoplasmic Source Analysis of Sterile Lines from 5460 Three-line Hybrid Rice Varieties [J]. Chinese Journal OF Rice Science, 2024, 38(6): 653-664. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||