
Chinese Journal OF Rice Science ›› 2018, Vol. 32 ›› Issue (4): 325-334.DOI: 10.16819/j.1001-7216.2018.7122
• Orginal Article • Previous Articles Next Articles
Kunneng ZHOU, Jiafa XIA, Tingchen MA, Yuanlei WANG, Zefu LI*( )
)
Received:2017-09-28
Revised:2018-01-14
Online:2018-07-10
Published:2018-07-10
Contact:
Zefu LI
通讯作者:
李泽福
基金资助:CLC Number:
Kunneng ZHOU, Jiafa XIA, Tingchen MA, Yuanlei WANG, Zefu LI. Mapping and Mutation Analysis of Stripe Leaf and White Panicle Gene SLWP in Rice[J]. Chinese Journal OF Rice Science, 2018, 32(4): 325-334.
周坤能, 夏加发, 马廷臣, 王元垒, 李泽福. 水稻条纹叶和白穗基因SLWP的定位及变异分析[J]. 中国水稻科学, 2018, 32(4): 325-334.
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.ricesci.cn/EN/10.16819/j.1001-7216.2018.7122
| 引物用途与名称 Usage and name | 正向引物 Forward sequence(5′-3′) | 反向引物 Reverse sequence(5′-3′) |
|---|---|---|
| 定位和测序引物 Primers for mapping and sequencing | ||
| C6-4 | CAGTTAACACCAATCCAATCCA | CCAAATGGGCAGTAGTTTGAA |
| C6-5 | GCTTCTCCCGGAGTATGTCA | TGGTCTGAAAAGTGCCAAAA |
| C6-6 | CCCCAAGCTTGTTTGTCTCT | TTGGGTCTTTGAGCTTCCTC |
| N4 | TCAAGTTGCTAAACCTTATCTG | AGATGAACTGTGCTAAAAGATG |
| N12 | GTAACTTAAAAGCCAATGTTGA | GAGTACTACCATCCATCCCTGT |
| N14 | TCTGTGGACGTAGTAGGTTGA | CCTTCCTTAGGTCTGGCTC |
| slwp-g | ACAACCCCAAATCCCCATCCA | ACCACGCGCATGTATTACTACT |
| 定量PCR引物 Primers for quantitative RT-PCR | ||
| PORA | ATCACCAAGGGCTACGTCTC | GAGTTGTTGTTCCAGCTCCA |
| HEMA1 | CACCAGTCTGAATCATAT | CTACCACTTCTCTAATCC |
| YGL1 | TGGACAGTTGAAGATGTT | GAATAGGACGGTAAGGTT |
| CHLI | AGTAACCTTGGTGCTGTG | AATCCATCAACATTCAACTCTG |
| CHLH | CTATACATTCGCCACACT | TATCACACAACTCCCAAG |
| CHLD | GGAAAGAGAGGGCATTAG | CAATACGATCAAGTAAGTGTT |
| FtsZ | GTTGGTGTTTCTTCCAGCAA | CCTCAATAGACGACCCGATT |
| RpoTp2 | AAGTCTGGCTTACGCTGGTT | AGGATCCTCAGCATTCATCC |
| rpoA | AAATCGTTGATACGGCACAA | ATTCACATTTCGAACAGGCA |
| rpoB | GCATTGTTGGAACTGGATTG | GCCGATGGGTAACTAAAGGA |
| rbcL | GTTGAAAGGGATAAGTTGA | AATGGTTGTGAGTTTACG |
| rbcS | TCATCAGCTTCATCGCCTAC | ACTGGGAACACACGAAACAA |
| psbA | AAGTTTCTCTGATGGTATG | ATAGCACTGAATAGGGAA |
| psaA | GAGATACCACTTCCTCAT | ACTAAGAAATTCTGCGTATT |
| psaB | TTGGTATTGCTACCGCACAT | CCGGACGTCCATAGAAAGAT |
| psbB | TCATATTGCTGCGGGTACAT | AGTTGCTGACCCATACCACA |
| psbC | TACAACCTTGGCAAGAACGA | TACGCCACCCACAGAATTTA |
| SLWP | AGTAATACATGCGCGTGGTG | ACACGGGCAGCTGATACTAA |
| RNRS2 | AGATGTACAACGTCGCCAAC | GACATTACGGACGCCTTCTG |
| Ubq | GCTCCGTGGCGGTATCAT | CGGCAGTTGACAGCCCTAG |
Table 1 Primer sequences used in the study.
| 引物用途与名称 Usage and name | 正向引物 Forward sequence(5′-3′) | 反向引物 Reverse sequence(5′-3′) |
|---|---|---|
| 定位和测序引物 Primers for mapping and sequencing | ||
| C6-4 | CAGTTAACACCAATCCAATCCA | CCAAATGGGCAGTAGTTTGAA |
| C6-5 | GCTTCTCCCGGAGTATGTCA | TGGTCTGAAAAGTGCCAAAA |
| C6-6 | CCCCAAGCTTGTTTGTCTCT | TTGGGTCTTTGAGCTTCCTC |
| N4 | TCAAGTTGCTAAACCTTATCTG | AGATGAACTGTGCTAAAAGATG |
| N12 | GTAACTTAAAAGCCAATGTTGA | GAGTACTACCATCCATCCCTGT |
| N14 | TCTGTGGACGTAGTAGGTTGA | CCTTCCTTAGGTCTGGCTC |
| slwp-g | ACAACCCCAAATCCCCATCCA | ACCACGCGCATGTATTACTACT |
| 定量PCR引物 Primers for quantitative RT-PCR | ||
| PORA | ATCACCAAGGGCTACGTCTC | GAGTTGTTGTTCCAGCTCCA |
| HEMA1 | CACCAGTCTGAATCATAT | CTACCACTTCTCTAATCC |
| YGL1 | TGGACAGTTGAAGATGTT | GAATAGGACGGTAAGGTT |
| CHLI | AGTAACCTTGGTGCTGTG | AATCCATCAACATTCAACTCTG |
| CHLH | CTATACATTCGCCACACT | TATCACACAACTCCCAAG |
| CHLD | GGAAAGAGAGGGCATTAG | CAATACGATCAAGTAAGTGTT |
| FtsZ | GTTGGTGTTTCTTCCAGCAA | CCTCAATAGACGACCCGATT |
| RpoTp2 | AAGTCTGGCTTACGCTGGTT | AGGATCCTCAGCATTCATCC |
| rpoA | AAATCGTTGATACGGCACAA | ATTCACATTTCGAACAGGCA |
| rpoB | GCATTGTTGGAACTGGATTG | GCCGATGGGTAACTAAAGGA |
| rbcL | GTTGAAAGGGATAAGTTGA | AATGGTTGTGAGTTTACG |
| rbcS | TCATCAGCTTCATCGCCTAC | ACTGGGAACACACGAAACAA |
| psbA | AAGTTTCTCTGATGGTATG | ATAGCACTGAATAGGGAA |
| psaA | GAGATACCACTTCCTCAT | ACTAAGAAATTCTGCGTATT |
| psaB | TTGGTATTGCTACCGCACAT | CCGGACGTCCATAGAAAGAT |
| psbB | TCATATTGCTGCGGGTACAT | AGTTGCTGACCCATACCACA |
| psbC | TACAACCTTGGCAAGAACGA | TACGCCACCCACAGAATTTA |
| SLWP | AGTAATACATGCGCGTGGTG | ACACGGGCAGCTGATACTAA |
| RNRS2 | AGATGTACAACGTCGCCAAC | GACATTACGGACGCCTTCTG |
| Ubq | GCTCCGTGGCGGTATCAT | CGGCAGTTGACAGCCCTAG |
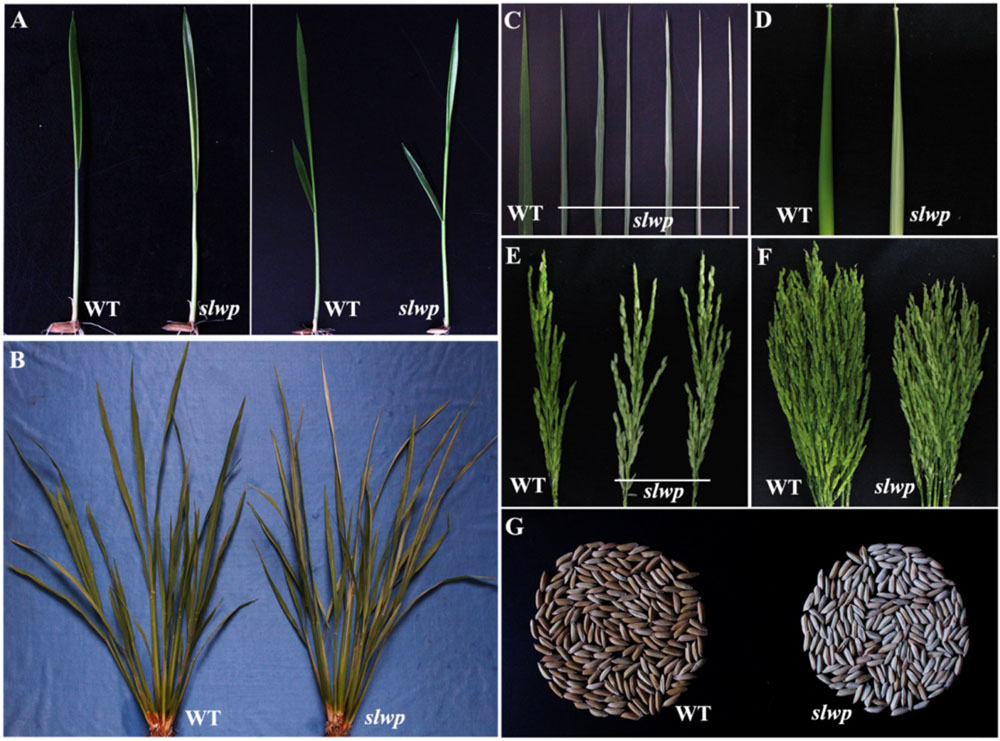
Fig. 1. Phenotypic characteristics of the slwp mutant and its wild type(WT). A, Phenotypes of two- and three-leaf stages; B and C, Leaf phenotype of tillering stage; D, Sheath phenotype of booting stage; E and F—Panicle phenotype of heading stage; G, Seed phenotype.
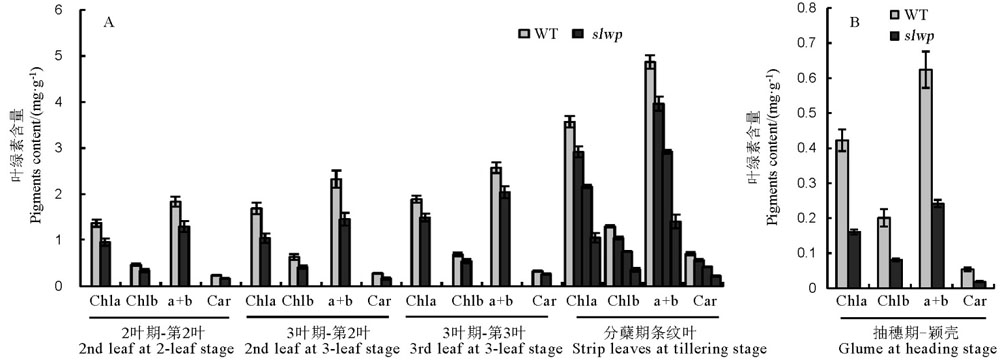
Fig. 2. Pigments determination of the slwp mutant and its wild type(WT). Chla, Chlorophyll a; chlb, Chlorophyll b; a+b, Total chlorophyll; Car―Carotene.
| 农艺性状 Agronomic trait | 野生型 Wild type | 突变体 swlp |
|---|---|---|
| 株高Plant height / cm | 124.7±3.4 | 98.2±4.8** |
| 播始历期Days from sowing to heading / d | 98.4±1.2 | 108.3±1.0** |
| 穗长Panicle length / cm | 25.5±1.1 | 22.1±1.3** |
| 有效穗数Number of effective panicles per plant | 7.1±1.4 | 6.6±1.7 |
| 每穗粒数Grain number per panicle | 245.1±32.6 | 145.0±21.0** |
| 结实率Seed-setting rate / % | 88.2±1.3 | 87.9±1.6 |
| 千粒重1000-grain weight / g | 30.8±0.4 | 27.9±0.6** |
| 理论产量Theoretical yield per plot / kg | 10.74±0.23 | 5.34±0.35** |
| 实际产量Actual yield per plot / kg | 9.16±0.26 | 4.43±0.47** |
Table 2 Agronomic traits of the wild type (WT) and slwp mutant.
| 农艺性状 Agronomic trait | 野生型 Wild type | 突变体 swlp |
|---|---|---|
| 株高Plant height / cm | 124.7±3.4 | 98.2±4.8** |
| 播始历期Days from sowing to heading / d | 98.4±1.2 | 108.3±1.0** |
| 穗长Panicle length / cm | 25.5±1.1 | 22.1±1.3** |
| 有效穗数Number of effective panicles per plant | 7.1±1.4 | 6.6±1.7 |
| 每穗粒数Grain number per panicle | 245.1±32.6 | 145.0±21.0** |
| 结实率Seed-setting rate / % | 88.2±1.3 | 87.9±1.6 |
| 千粒重1000-grain weight / g | 30.8±0.4 | 27.9±0.6** |
| 理论产量Theoretical yield per plot / kg | 10.74±0.23 | 5.34±0.35** |
| 实际产量Actual yield per plot / kg | 9.16±0.26 | 4.43±0.47** |
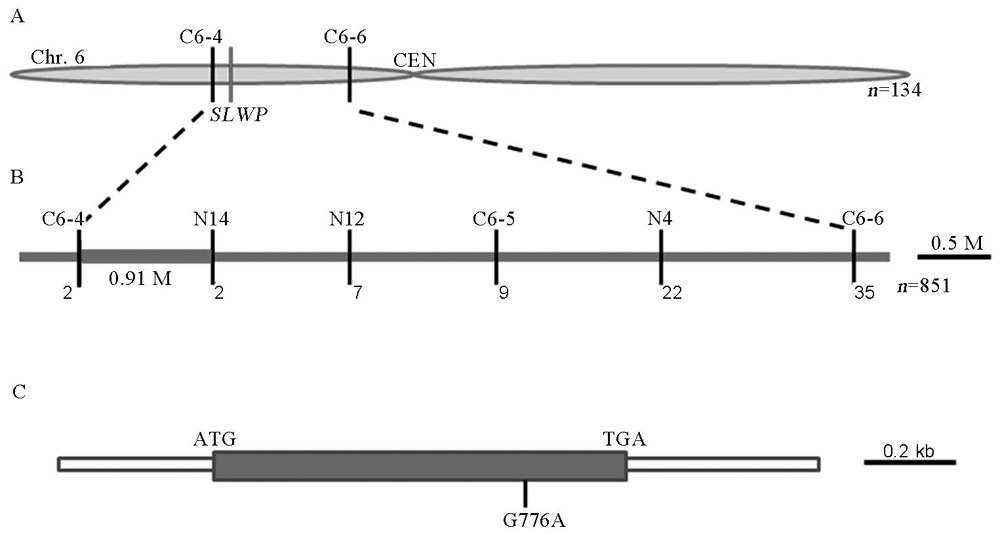
Fig. 3. Map-based cloning of the SLWP gene. A, Initial mapping of the SLWP gene; B, Fine mapping of the SLWP gene; C, Structure and mutant site of the SLWP gene; ATG and TGA indicate initiation codon and termination codon, respectively.
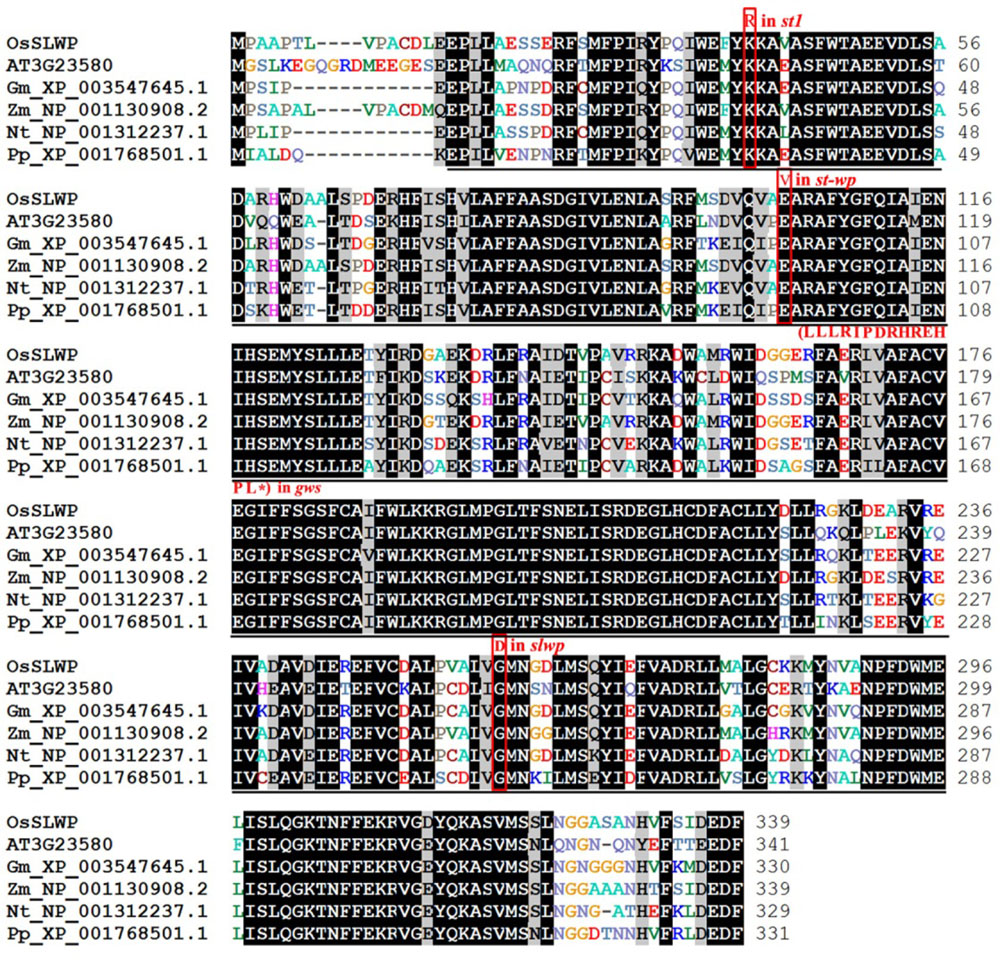
Fig. 4. Alignment of SLWP and SLWP-related proteins of different organisms. Sequences are for Oryza sativa LOC_Os06g14620(SLWP), Arabidopsis thaliana At3g23580, Glycine max XP_003547645.1, Zea mays NP_001130908.2, Nicotiana tabacum NP_001312237.1 and Physcomitrella patens XP_001768501.1; Underline represents nucleotide reductase domain; Red font and frame indicate mutant sites of ST1, STWP, GWS and SLWP.
| 突变体 Mutant | 野生型 Wild type | 氨基酸突变位点 Mutant site of amino acid | 叶片表型 Leaf phenotype | 穗部表型 Panicle phenotype | 主要农艺性状变化 Agronomic trait change compared with its wild type |
|---|---|---|---|---|---|
| st1 | FL176 | p. Lys40Arg | 4或5叶期至分蘖期出现条纹;抽穗后表型恢复Display stripe from 4- or 5-leaf stage to tillering, then revert to normal after heading | 无 None | 无 None |
| gws | 日本晴 Nipponbare | 104原有+插入14个氨基酸 104 original+Inserted 14 amino acid residues | 苗期至抽穗期叶片均出现条纹Display stripe from the seedling to heading stage | 无 None | 株高、穗长、有效分蘖数、结实率、千粒重、单株生物量均降低Decreased plant height, panicle length, tiller number, seed-setting rate, 1000-grain weight, and plant biomass per plant |
| st-wp | 9311 | p. Glu103Val | 苗期至抽穗期叶片均出现条纹Display stripe from the seedling to heading stage | 白穗 White panicle | 株高、穗长、每穗粒数、结实率、千粒重降低,抽穗期延长,有效穗数不变 Decreased plant height, panicle length, grain number per panicle, seed-setting rate, and 1000-grain weight; prolonged heading date; similar tiller number |
| slwp | 9311 | p. Gly259Asp | 苗期至抽穗期叶片出现条纹 Display stripe from the seedling to heading stage | 白穗 White panicle | 株高、穗长、每穗粒数、千粒重、理论产量、实际产量下降,抽穗期延长,有效穗数和结实率不变 Decreased plant height, panicle length, grain number per panicle, 1000-grain weight, theoretical yield, and actual yield; prolonged heading date, similar tiller number and seed-setting rate |
Table 3 Mutation analysis of SLWP.
| 突变体 Mutant | 野生型 Wild type | 氨基酸突变位点 Mutant site of amino acid | 叶片表型 Leaf phenotype | 穗部表型 Panicle phenotype | 主要农艺性状变化 Agronomic trait change compared with its wild type |
|---|---|---|---|---|---|
| st1 | FL176 | p. Lys40Arg | 4或5叶期至分蘖期出现条纹;抽穗后表型恢复Display stripe from 4- or 5-leaf stage to tillering, then revert to normal after heading | 无 None | 无 None |
| gws | 日本晴 Nipponbare | 104原有+插入14个氨基酸 104 original+Inserted 14 amino acid residues | 苗期至抽穗期叶片均出现条纹Display stripe from the seedling to heading stage | 无 None | 株高、穗长、有效分蘖数、结实率、千粒重、单株生物量均降低Decreased plant height, panicle length, tiller number, seed-setting rate, 1000-grain weight, and plant biomass per plant |
| st-wp | 9311 | p. Glu103Val | 苗期至抽穗期叶片均出现条纹Display stripe from the seedling to heading stage | 白穗 White panicle | 株高、穗长、每穗粒数、结实率、千粒重降低,抽穗期延长,有效穗数不变 Decreased plant height, panicle length, grain number per panicle, seed-setting rate, and 1000-grain weight; prolonged heading date; similar tiller number |
| slwp | 9311 | p. Gly259Asp | 苗期至抽穗期叶片出现条纹 Display stripe from the seedling to heading stage | 白穗 White panicle | 株高、穗长、每穗粒数、千粒重、理论产量、实际产量下降,抽穗期延长,有效穗数和结实率不变 Decreased plant height, panicle length, grain number per panicle, 1000-grain weight, theoretical yield, and actual yield; prolonged heading date, similar tiller number and seed-setting rate |
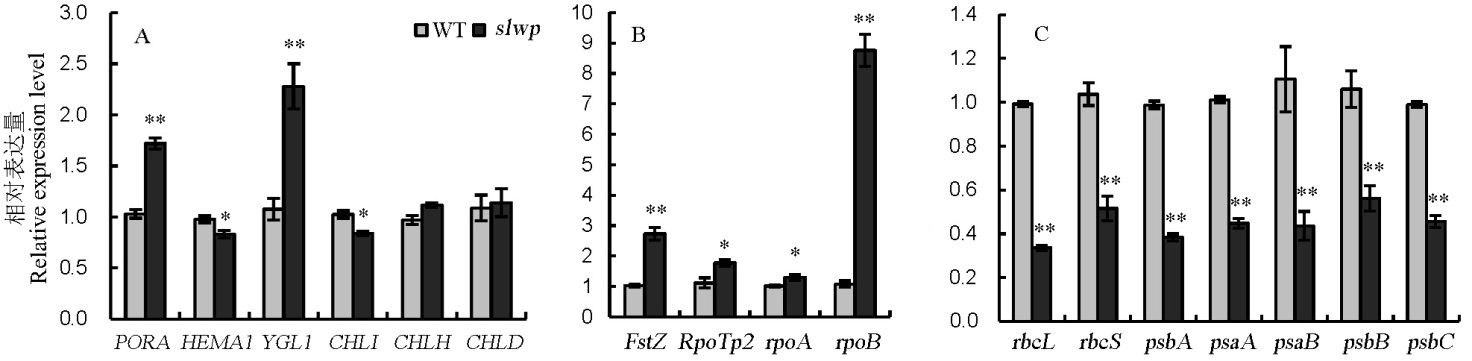
Fig. 6. Expression analysis of genes involved in chlorophyll biosynthesis, chloroplast development and photosynthesis between wild type(WT) and mutant. A, Expression analysis of genes associated with chlorophyll biosynthesis; B, Expression analysis of genes associated with chloroplast development; C, Expression analysis of genes associated with photosynthesis. * and ** represent significant difference at 5% and 1% levels, respectively.
| [1] | Fromme P, Melkozernov A, Jordan P, Krauss N.Structure and function of photosystem I: interaction with its soluble electron carriers and external antenna systems.FEBS Lett, 2003, 555: 40-44. |
| [2] | Lokstein H, Tian L, Polle J E W, DellaPenna D. Xanthophyll biosynthetic mutants of Arabidopsis thaliana: Altered nonphotochemical quenching of chlorophyll fluorescence is due to changes in Photosystem II antenna size and stability.Biochim Biophys Acta, 2002, 1553: 309-319. |
| [3] | Lv M Z, Chao D Y, Shan J X, Zhu M Z, Shi M, Gao J P, Lin H X.Rice carotenoid b-ring hydroxylase CYP97A4 is involved in lutein biosynthesis.Plant Cell Physiol, 2012, 53: 987-1002. |
| [4] | Nagata N, Tanaka R, Satoh S, Tanaka A.Identification of a vinyl reductase gene for chlorophyll synthesis in Arabidopsis thaliana and implications for the evolution of Prochlorococcus species. Plant Cell, 2005, 17(1): 233-240. |
| [5] | Yoo S C, Cho S H, Sugimoto H, Li J, Kusumi K, Koh H J, Iba K, Paek N C.Rice Virescent3 and Stripe1 encoding the large and small subunits of ribonucleotide reductase are required for chloroplast biogenesis during early leaf development. Plant Physiol, 2009, 150: 388-401. |
| [6] | Kusumi K, Mizutani A, Nishimura M, Iba K.A virescent gene V1 determines the expression timing of plastid genes for transcription/translation apparatus during early leaf development in rice. Plant J, 1997, 12: 1241-1250. |
| [7] | Kusumi K, Sakata C, Nakamura T, Kawasaki S, Yoshimura A, Iba K.A plastid protein NUS1 is essential for build-up of the genetic system for early chloroplast development under cold stress conditions.Plant J, 2011, 68: 1039-1050. |
| [8] | Sugimoto H, Kusumi K, Tozawa Y, Yazaki J, Kishimoto N, Kikuchi S, Iba K.The virescent-2 mutation inhibits translation of plastid transcripts for the plastid genetic system at an early stage of chloroplast differentiation. Plant Cell Physiol, 2004, 45: 985-996. |
| [9] | Sugimoto H, Kusumi K, Noguchi K, Yano M, Yoshimura A, Iba K.The rice nuclear gene, VIRESCENT 2 , is essential for chloroplast development and encodes a novel type of guanylate kinase targeted to plastids and mitochondria. Plant J, 2007, 52: 512-527. |
| [10] | Dong H, Fei G L, Wu C Y, Wu F Q, Sun Y Y, Chen M J, Ren Y L, Zhou K N, Cheng Z J, Wang J L, Jiang L, Zhang X, Guo X P, Lei C L, Su N, Wang H Y, Wan J M. A rice virescent-yellow leaf mutant reveals new insights into the role and assembly of plastid caseinolytic protease in higher plants. Plant Physiol, 2013, 162: 1867-1880. |
| [11] | Tan J, Tan Z, Wu F, Sheng P, Heng Y, Wang X, Ren Y, Wang J, Guo X, Zhang X, Cheng Z, Jiang L, Liu X, Wang H, Wan J.A novel chloroplast-localized pentatricopeptide repeat protein involved in splicing affects chloroplast development and abiotic stress response in rice.Mol Plant, 2014, 7: 1329-1349. |
| [12] | Wang Y, Ren Y, Zhou K, Liu L, Wang J, Xu Y, Zhang H, Zhang L, Feng Z, Wang L, Ma W, Wang Y, Guo X, Zhang X, Lei C, Cheng Z, Wan J.WHITE STRIPE LEAF4 encodes a novel P-type PPR protein required for chloroplast biogenesis during early leaf development. Front Plant Sci, 2017, 8: 1116. |
| [13] | Wang L, Wang C, Wang Y, Niu M, Ren Y, Zhou K, Zhang H, Lin Q, Wu F, Cheng Z, Wang J, Zhang X, Guo X, Jiang L, Lei C, Wang J, Zhu S, Zhao Z, Wan J.WSL3, a component of the plastid-encoded plastid RNA polymerase, is essential for early chloroplast development in rice.Plant Mol Biol, 2016, 92: 581-595. |
| [14] | Zhou K N, Ren Y L, Zhou F, Wang Y, Zhang L, Lyu J, Wang Y H, Zhao S L, Ma W W, Zhang H, Wang L W, Wang C M, Wu F Q, Zhang X, Guo X P, Cheng Z J, Wang J L, Lei C L, Jiang L, Li Z F, Wan J M.Young Seedling Stripe1 encodes a chloroplast nucleoid- associated protein required for chloroplast development in rice seedlings. Planta, 2017, 245: 45-60. |
| [15] | Sheng P, Tan J, Jin M, Wu F, Zhou K, Ma W, Heng Y, Wang J, Guo X, Zhang X, Cheng Z, Liu L, Wang C, Liu X, Wan J.Albino midrib 1 , encoding a putative potassium efflux antiporter, affects chloroplast development and drought tolerance in rice. Plant Cell Rep, 2014, 33: 1581-1594. |
| [16] | 李红昌, 钱前, 王赟, 李晓波, 朱立煌, 徐吉臣. 水稻白穗突变体基因的鉴定和染色体定位. 科学通报, 2003, 48(3): 268-270. |
| Li H C, Qian Q, Wang Y, Li X B, Zhu L H, Xu J C.Identification and gene mapping of white panicle mutant in rice.Chin Sci Bull, 2003, 48(3): 268-270. (in Chinese with English abstract) | |
| [17] | 金怡, 刘合芹, 汪得凯, 陶跃之. 一个水稻苗期白条纹及抽穗期白穗突变体的鉴定和基因定位. 中国水稻科学, 2011, 25(5): 461-466. |
| Jin Y, Liu H Q, Wang D K, Tao Y Z.Genetic analysis and gene mapping of a white striped leaf and white panicle mutant in rice.Chin J Rice Sci, 2011, 25(5): 461-466. (in Chinese with English abstract) | |
| [18] | 王晓雯, 蒋钰东, 廖红香, 杨波, 邹帅宇, 朱小燕, 何光华, 桑贤春. 水稻白穗突变体wp4的鉴定与基因精细定位. 作物学报, 2015, 41(6): 838-844. |
| Wang X W, Jiang Y D, Niao H X, Yang B, Zou S Y, Zhu X Y, He G H, Sang X C.Identification and gene mapping of white panicle mutant wp4 in Oryza sativa. Acta Agron Sin, 2015, 41(6): 838-844. (in Chinese with English abstract) | |
| [19] | Song J, Wei X J, Shao G N, Sheng Z H, Cheng D, Liu C, Jiao G A, Xie L H, Tang S Q, Hu P S.The rice nuclear gene WLP1 encoding a chloroplast ribosome L13 protein is needed for chloroplast development in rice grown under low temperature conditions. Plant Mol Biol, 2014, 84: 301-314. |
| [20] | Wang Y L, Wang C M, Zheng M, Lyu J, Xu Y, Li X H, Niu M, Long W H, Wang D, Wang H Y, Terzaghi W, Wang Y H, Wan J M.WHITE PANICLE1 , a Val-tRNA synthetase regulating chloroplast ribosome biogenesis in rice, is essential for early chloroplast development. Plant Physiol, 2016, 170: 2120-2123. |
| [21] | Zhou K N, Xia J F, Wang Y L, Ma T C, Li Z F.A Young Seedling Stripe2 phenotype in rice is caused by mutation of a chloroplast-localized nucleoside diphosphate kinase 2 required for chloroplast biogenesis. Genet Mol Biol, 2017, 40: 630-642. |
| [22] | 许凤华, 程治军, 王久林, 吴自明, 孙伟, 张欣, 雷财林, 王洁, 吴赴清, 郭秀平, 刘玲珑, 万建民. 水稻白条纹叶Gws基因的精细定位与遗传分析. 作物学报, 2010, 36(5): 713-720. |
| Xu F H, Cheng Z J, Wang J L, Wu Z M, Sun W, Zhang X, Lei C L, Wang J, Wu F Q, Guo X P, Liu L L, Wan J M.Genetic analysis and fine-mapping of Gws gene using green-white-stripe rice mutant. Acta Agron Sin, 2010, 36(5): 713-720. (in Chinese with English abstract) | |
| [23] | 陈叶平, 翟哲, 杨文君, 孙健, 舒小丽, 吴殿星. 水稻条白叶和白穗突变基因St-wp的遗传分析与精细定位. 核农学报, 2015, 29(7): 1246-1252. |
| Chen Y P, Zhai Z, Yang W J, Sun J, Shu X L, Wu D X.Genetic analysis and fine mapping of St-wp gene in mutant rice with stripe white leaf and white panicle. Acta Agric Nucl Sin, 2015, 29(7): 1246-1252. (in Chinese with English abstract) | |
| [24] | Eriksson M, Uhlin U, Ramaswamy S, Ekberg M, Regnström K, Sjöberg B M, Eklund H.Binding of allosteric effectors to ribonucleotide reductase protein R1: reduction of active-site cysteines promotes substrate binding.Structure, 1997, 5: 1077-1092. |
| [1] | GAO Junru, QUAN Hongyu, YUAN Liuzhen, LI Qinying, QIAO Lei, LI Wenqiang. Map-based Cloning and Functional Analysis of a New Allele of D1, a Gene Controlling Plant Height in Rice (Oryza sativa L.) [J]. Chinese Journal OF Rice Science, 2024, 38(2): 140-149. |
| [2] | XU Huan, ZHOU Tao, SUN Yue, WANG Mumei, YANG Yachun, MA Hui, LI Hao, XU Dawei, ZHOU Hai, YANG Jianbo, NI Jinlong. Characterization and Gene Mapping of a Glume Lesion Mimic Mutant glmm1 in Rice [J]. Chinese Journal OF Rice Science, 2023, 37(5): 497-506. |
| [3] | SUN Zhiguang, DAI Huimin, CHEN Tingmu, LI Jingfang, CHI Ming, ZHOU Zhenling, LIU Yan, LIU Jinbo, XU Bo, XING Yungao, YANG Bo, LI Jian, LU Baiguan, FANG Zhaowei, WANG Baoxiang, XU Dayong. Phenotypic Identification and Gene Mapping of a Lesion Mimic Mutant lmm7 in Rice [J]. Chinese Journal OF Rice Science, 2022, 36(4): 357-366. |
| [4] | YAO Shu, ZHANG Yadong, LU Kai, WANG Cailin. Progress in Functions, Allelic Variations and Interactions of Soluble Starch Synthases Genes SSⅡa and SSⅢa in Rice [J]. Chinese Journal OF Rice Science, 2022, 36(3): 227-236. |
| [5] | QIU Linlin, LIU Qiao, FU Yaping, LIU Wenzhen, HU Guocheng, ZHAI Yufeng, PANG Bo, WANG Dekai. Identification and Gene Cloning of DSP2 in Rice (Oryza sativa L.) [J]. Chinese Journal OF Rice Science, 2022, 36(2): 150-158. |
| [6] | Yuxiang LI, Hairong LIN, Qian LIANG, Guodong WANG. Effects of Dopamine Priming on Seed Germination and Seedling Growth of Rice Under Salt Stress [J]. Chinese Journal OF Rice Science, 2021, 35(5): 487-494. |
| [7] | Chen WANG, Beifang WANG, Yingxin ZHANG, Yongrun CAO, Yue ZHANG, Min JIANG, Kangji BIAN, Xiaohui ZHANG, Qunen LIU. Identification and Gene Mapping of a Lesion Mimic Mutant lm8015-2 in Rice [J]. Chinese Journal OF Rice Science, 2021, 35(4): 352-358. |
| [8] | Ke LI, Qing YU, Yunji XU, Jianchang YANG. Research Progress in Agronomic and Physiological Traits of Early Leaf Senescence Mutants in Rice [J]. Chinese Journal OF Rice Science, 2020, 34(2): 104-114. |
| [9] | Shilin DING, Chaolei LIU, Qian QIAN, Zhenyu GAO. Research Advances on Molecular Genetic Mechanism for Cadmium Absorption and Transportation in Rice [J]. Chinese Journal OF Rice Science, 2019, 33(5): 383-390. |
| [10] | Feng LÜ, Qianying TANG, Qiming WANG, Hai ZHENG, Shimin YOU, Wenting BAI, Yanjia XIAO, Zhigang ZHAO, Jianmin WAN. Map-based Cloning of Female Abortion (FA) Gene in Rice [J]. Chinese Journal OF Rice Science, 2018, 32(6): 519-528. |
| [11] | Hua ZHAO, Fubiao WANG, Zhanyu HAN, zakari SHAMSU-ADO, Zhaowei LI, Gang PAN, Fangmin CHENG. Senescence-specific Changes in Endogenous IAA Content and Its Conjugated Status in Rice Flag Leaf as Affected by Nitrogen Level [J]. Chinese Journal OF Rice Science, 2018, 32(6): 591-600. |
| [12] | Shaofeng JIANG, Chenjiaozi WANG, Canwei SHU, Erxun ZHOU. Cloning and Expression Analysis of RsPhmGene inRhizoctoniasolani AG-1ⅠA of Rice Sheath Blight Pathogen [J]. Chinese Journal OF Rice Science, 2018, 1(1): 111-118. |
| [13] | Hong WANG, Yingxin ZHANG, Lianping SUN, Shuai MENG, Peng XU, Weixun WU, Shihua CHENG, Liyong CAO, Xihong SHEN. Map-Based Cloning of OsCAD2 Regulating Golden Hull and Internode in Rice [J]. Chinese Journal OF Rice Science, 2017, 31(5): 465-474. |
| [14] | Liting SUN, Tianzi LIN, Yunlong WANG, Mei NIU, Tingting HU, Shijia LIU, Yihua WANG, Jianmin WAN. Phenotypic Analysis and Gene Mapping of a White Stripe Mutant st13 in Rice [J]. Chinese Journal OF Rice Science, 2017, 31(4): 355-363. |
| [15] | Zuopeng XU, Chongyuan ZHONG, Lijia ZHANG, Qiaoquan LIU. Identification and Gene Cloning of the Brittle Culm Mutant bc1-wu3 in Rice [J]. Chinese Journal OF Rice Science, 2017, 31(2): 157-165. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||