
Chinese Journal OF Rice Science ›› 2018, Vol. 32 ›› Issue (3): 207-218.DOI: 10.16819/j.1001-7216.2018.7120
• Research Papers • Next Articles
Mengyu ZHOU, Xinwei SONG, Jing XU, Xue FU, Ting LI, Yuchen ZHU, Xingyun XIAO, Yijian MAO, Dali ZENG, Jiang HU, Li ZHU, Deyong REN, Zhenyu GAO, Longbiao GUO, Qian QIAN, Mingguo WU, Jianrong LIN*( ), Guangheng ZHANG*(
), Guangheng ZHANG*( )
)
Received:2017-09-27
Revised:2018-01-23
Online:2018-05-10
Published:2018-05-10
Contact:
Jianrong LIN, Guangheng ZHANG
周梦玉, 宋昕蔚, 徐静, 付雪, 李婷, 朱雨晨, 肖幸运, 毛一剑, 曾大力, 胡江, 朱丽, 任德勇, 高振宇, 郭龙彪, 钱前, 吴明国, 林建荣*( ), 张光恒*(
), 张光恒*( )
)
通讯作者:
林建荣,张光恒
基金资助:CLC Number:
Mengyu ZHOU, Xinwei SONG, Jing XU, Xue FU, Ting LI, Yuchen ZHU, Xingyun XIAO, Yijian MAO, Dali ZENG, Jiang HU, Li ZHU, Deyong REN, Zhenyu GAO, Longbiao GUO, Qian QIAN, Mingguo WU, Jianrong LIN, Guangheng ZHANG. Construction of Genetic Map andMapping andVerification of Grain TraitsQTLs Using Recombinant Inbred LinesDerived from a Cross BetweenindicaC84 andjaponicaCJ16B[J]. Chinese Journal OF Rice Science, 2018, 32(3): 207-218.
周梦玉, 宋昕蔚, 徐静, 付雪, 李婷, 朱雨晨, 肖幸运, 毛一剑, 曾大力, 胡江, 朱丽, 任德勇, 高振宇, 郭龙彪, 钱前, 吴明国, 林建荣, 张光恒. 籼稻C84和粳稻春江16B重组自交系遗传图谱构建及籽粒性状QTL定位与验证[J]. 中国水稻科学, 2018, 32(3): 207-218.
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.ricesci.cn/EN/10.16819/j.1001-7216.2018.7120
| 性状 Trait | 地点 Site | 亲本Parent | 重组自交系群体RIL population | ||||
|---|---|---|---|---|---|---|---|
| C84 | 春江16BCJ16B | 平均Mean | 范围Range | ||||
| 粒长 Grain length / mm | 杭州 Hangzhou | 8.2±0.2 | 7.4±0.2** | 7.74 | 6.4~9.0 | ||
| 陵水Lingshui | 7.8±0.1 | 7.0±0.1** | 7.56 | 6.5~8.7 | |||
| 粒宽Grain width / mm | 杭州 Hangzhou | 2.9±0.1 | 3.2±0.0** | 3.11 | 2.6~3.8 | ||
| 陵水Lingshui | 3.1±0.1 | 3.2±0.1* | 3.22 | 2.6~3.7 | |||
| 粒厚Grain thickness / mm | 杭州 Hangzhou | 1.98±0.01 | 2.26±0.04** | 2.06 | 1.79~2.38 | ||
| 陵水Lingshui | 2.03±0.04 | 2.28±0.04** | 2.08 | 1.75~2.34 | |||
| 长宽比Lengthtowidth ratio | 杭州 Hangzhou | 2.8±0.1 | 2.3±0.1* | 2.52 | 2.0~3.4 | ||
| 陵水Lingshui | 2.5±0.3 | 2.2±0.1** | 2.37 | 1.9~3.0 | |||
| 千粒重1000-grain weight / g | 杭州 Hangzhou | 23.7±0.4 | 25.1±0.3* | 22.17 | 13.8~33.2 | ||
| 陵水Lingshui | 24.1±0.1 | 25.2±0.1* | 22.98 | 13.3~30.8 | |||
Table 1 Grain length, width, thickness, length-to-width ratio and 1000-grain weight inC84 and CJ16Band their recombinant inbred line population in Hangzhou, Zhejiang Province and Lingshui, Hainan Province, China in 2016.
| 性状 Trait | 地点 Site | 亲本Parent | 重组自交系群体RIL population | ||||
|---|---|---|---|---|---|---|---|
| C84 | 春江16BCJ16B | 平均Mean | 范围Range | ||||
| 粒长 Grain length / mm | 杭州 Hangzhou | 8.2±0.2 | 7.4±0.2** | 7.74 | 6.4~9.0 | ||
| 陵水Lingshui | 7.8±0.1 | 7.0±0.1** | 7.56 | 6.5~8.7 | |||
| 粒宽Grain width / mm | 杭州 Hangzhou | 2.9±0.1 | 3.2±0.0** | 3.11 | 2.6~3.8 | ||
| 陵水Lingshui | 3.1±0.1 | 3.2±0.1* | 3.22 | 2.6~3.7 | |||
| 粒厚Grain thickness / mm | 杭州 Hangzhou | 1.98±0.01 | 2.26±0.04** | 2.06 | 1.79~2.38 | ||
| 陵水Lingshui | 2.03±0.04 | 2.28±0.04** | 2.08 | 1.75~2.34 | |||
| 长宽比Lengthtowidth ratio | 杭州 Hangzhou | 2.8±0.1 | 2.3±0.1* | 2.52 | 2.0~3.4 | ||
| 陵水Lingshui | 2.5±0.3 | 2.2±0.1** | 2.37 | 1.9~3.0 | |||
| 千粒重1000-grain weight / g | 杭州 Hangzhou | 23.7±0.4 | 25.1±0.3* | 22.17 | 13.8~33.2 | ||
| 陵水Lingshui | 24.1±0.1 | 25.2±0.1* | 22.98 | 13.3~30.8 | |||
| 地点Site 性状Trait | HZ-GL | HZ-GW | HZ-GT | HZ-LWR | HZ-TGW | LS-GL | LS-GW | LS-GT | LS-LWR | |
|---|---|---|---|---|---|---|---|---|---|---|
| 杭州 | 粒宽 GW | –0.104** | ||||||||
| Hangzhou | 粒厚 GT | 0.073** | 0.396** | |||||||
| (HZ) | 长宽比LWR | 0.672** | –0.801** | –0.248** | ||||||
| 千粒重TGW | 0.441** | 0.443** | 0.404** | –0.066** | ||||||
| 陵水 | 粒长 GL | 0.809** | ||||||||
| Lingshui | 粒宽 GW | 0.817** | –0.016** | |||||||
| (LS) | 粒厚 GT | 0.270** | 0.117** | 0.164** | ||||||
| 长宽比LWR | 0.854** | 0.660** | –0.758** | –0.030** | ||||||
| 千粒重 TGW | 0.529** | 0.438** | 0.508** | 0.287** | –0.094** | |||||
Table 2 Correlationship analysis on grain shape traits in C84/CJ16B recombinant inbred lines.
| 地点Site 性状Trait | HZ-GL | HZ-GW | HZ-GT | HZ-LWR | HZ-TGW | LS-GL | LS-GW | LS-GT | LS-LWR | |
|---|---|---|---|---|---|---|---|---|---|---|
| 杭州 | 粒宽 GW | –0.104** | ||||||||
| Hangzhou | 粒厚 GT | 0.073** | 0.396** | |||||||
| (HZ) | 长宽比LWR | 0.672** | –0.801** | –0.248** | ||||||
| 千粒重TGW | 0.441** | 0.443** | 0.404** | –0.066** | ||||||
| 陵水 | 粒长 GL | 0.809** | ||||||||
| Lingshui | 粒宽 GW | 0.817** | –0.016** | |||||||
| (LS) | 粒厚 GT | 0.270** | 0.117** | 0.164** | ||||||
| 长宽比LWR | 0.854** | 0.660** | –0.758** | –0.030** | ||||||
| 千粒重 TGW | 0.529** | 0.438** | 0.508** | 0.287** | –0.094** | |||||
| 染色体 Chromosome | 物理距离 Physical distance/Mb | 遗传距离 Genetic distance /cM | 平均物理距离 Mean physical distance /Mb | 平均遗传距离 Mean genetic distance /cM | 标记数 Number of markers |
|---|---|---|---|---|---|
| 1 | 46.15 | 192.8 | 2.71 | 11.34 | 17 |
| 2 | 35.93 | 128.0 | 2.57 | 9.14 | 14 |
| 3 | 36.41 | 145.9 | 2.28 | 9.12 | 16 |
| 4 | 35.28 | 148.5 | 1.96 | 8.25 | 18 |
| 5 | 29.89 | 106.9 | 1.99 | 7.13 | 15 |
| 6 | 31.25 | 104.8 | 2.84 | 9.53 | 11 |
| 7 | 29.69 | 128.5 | 2.12 | 9.18 | 14 |
| 8 | 28.44 | 107.2 | 2.59 | 9.75 | 11 |
| 9 | 23.01 | 77.5 | 2.30 | 7.75 | 10 |
| 10 | 23.13 | 72.5 | 2.57 | 8.06 | 9 |
| 11 | 28.51 | 115.5 | 2.38 | 9.63 | 12 |
| 12 | 27.49 | 100.3 | 2.50 | 9.12 | 11 |
| 合计 Total | 375.18 | 1428.4 | 2.40 | 9.04 | 158 |
Table 3 Distribution of the markers used in the study on 12chromosomes of rice.
| 染色体 Chromosome | 物理距离 Physical distance/Mb | 遗传距离 Genetic distance /cM | 平均物理距离 Mean physical distance /Mb | 平均遗传距离 Mean genetic distance /cM | 标记数 Number of markers |
|---|---|---|---|---|---|
| 1 | 46.15 | 192.8 | 2.71 | 11.34 | 17 |
| 2 | 35.93 | 128.0 | 2.57 | 9.14 | 14 |
| 3 | 36.41 | 145.9 | 2.28 | 9.12 | 16 |
| 4 | 35.28 | 148.5 | 1.96 | 8.25 | 18 |
| 5 | 29.89 | 106.9 | 1.99 | 7.13 | 15 |
| 6 | 31.25 | 104.8 | 2.84 | 9.53 | 11 |
| 7 | 29.69 | 128.5 | 2.12 | 9.18 | 14 |
| 8 | 28.44 | 107.2 | 2.59 | 9.75 | 11 |
| 9 | 23.01 | 77.5 | 2.30 | 7.75 | 10 |
| 10 | 23.13 | 72.5 | 2.57 | 8.06 | 9 |
| 11 | 28.51 | 115.5 | 2.38 | 9.63 | 12 |
| 12 | 27.49 | 100.3 | 2.50 | 9.12 | 11 |
| 合计 Total | 375.18 | 1428.4 | 2.40 | 9.04 | 158 |
| 性状 Trait | QTL | 标记区间 Marker interval | 位置 Position | 加性效应 Additive effect | 贡献率 EPV/% | 已克隆基因 Genecloned | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HZ | LS | HZ | LS | HZ | LS | |||||||||||
| 粒长 | qGL1 | RM3252 | –RM1247 | 2.0 | 0.0925 | 5.75 | ||||||||||
| Grain length | qGL2.1 | H2-2 | –H2-3 | 28.1 | –0.1539 | 9.01 | FUWA | |||||||||
| qGL2.2 | H2-5 | –H2-6 | 39.8 | –0.0804 | 7.05 | |||||||||||
| qGL5 | M17954R | –H5-7 | 32.5 | –0.1272 | 7.52 | JMJ703;SRS3/ OsKinesin-13A/sar1 | ||||||||||
| qGL7.1 | RM11 | –RM3826 | 81.0 | –0.1675 | 10.09 | |||||||||||
| qGL7.2 | RM3826 | –RM1132 | 90.7 | –0.1868 | 14.40 | |||||||||||
| qGL10 | RM6370 | –RM5689 | 14.0 | 18.0 | –0.1912 | –0.1794 | 11.97 | 10.33 | OsPCR1 | |||||||
| qGL11 | H11 | –4-H11-5 | 59.7 | 0.0768 | 5.89 | |||||||||||
| qGL12 | RM1337 | –RM3739 | 59.7 | 0.2160 | 11.04 | |||||||||||
| 粒宽 | qGW1 | H1-5 | –RM8097 | 141.6 | –0.0396 | 3.51 | ||||||||||
| Grain width | qGW2.1 | H2-6 | –H2-7 | 47.0 | 0.1134 | 9.58 | ||||||||||
| qGW2.2 | H2-7 | –RM262 | 52.7 | 0.0977 | 9.34 | |||||||||||
| qGW5 | RM17954 | –H5-7 | 27.5 | 23.5 | 0.1202 | 0.1143 | 12.28 | 11.44 | qGW5;JMJ703 | |||||||
| qGW6 | H6-2 | –RM3496 | 47.0 | 0.0728 | 7.06 | |||||||||||
| 粒厚 | qGT1 | RM8097 | –RM6703 | 163.5 | 163.5 | –0.1074 | –0.0907 | 12.75 | 12.59 | |||||||
| Grain thickness | qGT2.1 | H2-3 | –H2-4 | 33.2 | 0.0919 | 9.01 | ||||||||||
| qGT2.2 | H2-5 | –H2-6 | 41.8 | 0.0823 | 9.51 | |||||||||||
| qGT6 | wgwl | –RM3183 | 35.2 | 0.0708 | 8.21 | |||||||||||
| qGT9 | RM242 | –H9-6 | 69.3 | 0.0830 | 9.69 | |||||||||||
| 长宽比 | qGLW2.1 | H2-4 | –RM300 | 34.9 | –0.0787 | 13.88 | ||||||||||
| Length to width | qGLW2.2 | H2-5 | –H2-6 | 42.8 | –0.1332 | 15.61 | ||||||||||
| qGLW2 | H2-6 | –H2-7 | 50.0 | –0.0496 | 12.95 | |||||||||||
| qGLW5 | RM17954 | –H5-7 | 28.5 | 27.5 | –0.1598 | –0.1302 | 17.25 | 12.75 | ||||||||
| qGLW11 | RM167 | –H11-2 | 30.9 | 0.0519 | 7.79 | |||||||||||
| qGLW12 | RM1337 | –RM3739 | 51.7 | 0.0975 | 6.25 | |||||||||||
| 千粒重 | qTGW1.1 | H1-5 | –RM8097 | 144.6 | –1.1430 | 8.17 | OsTRBF3 | |||||||||
| 1000-grain | qTGW1.2 | RM8097 | –RM6703 | 148.5 | –1.0656 | 8.43 | ||||||||||
| weight | qTGW4 | RM317 | –RM5879 | 113.5 | 1.1454 | 7.41 | ||||||||||
| qTGW5 | RM18751 | –RM3476 | 85.1 | –1.2472 | 10.55 | |||||||||||
| qTGW9 | H9-4 | –RM3700 | 52.6 | –0.8719 | 4.55 | |||||||||||
Table 4 QTLs for grain shape-related traits of recombinant inbred lines.
| 性状 Trait | QTL | 标记区间 Marker interval | 位置 Position | 加性效应 Additive effect | 贡献率 EPV/% | 已克隆基因 Genecloned | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HZ | LS | HZ | LS | HZ | LS | |||||||||||
| 粒长 | qGL1 | RM3252 | –RM1247 | 2.0 | 0.0925 | 5.75 | ||||||||||
| Grain length | qGL2.1 | H2-2 | –H2-3 | 28.1 | –0.1539 | 9.01 | FUWA | |||||||||
| qGL2.2 | H2-5 | –H2-6 | 39.8 | –0.0804 | 7.05 | |||||||||||
| qGL5 | M17954R | –H5-7 | 32.5 | –0.1272 | 7.52 | JMJ703;SRS3/ OsKinesin-13A/sar1 | ||||||||||
| qGL7.1 | RM11 | –RM3826 | 81.0 | –0.1675 | 10.09 | |||||||||||
| qGL7.2 | RM3826 | –RM1132 | 90.7 | –0.1868 | 14.40 | |||||||||||
| qGL10 | RM6370 | –RM5689 | 14.0 | 18.0 | –0.1912 | –0.1794 | 11.97 | 10.33 | OsPCR1 | |||||||
| qGL11 | H11 | –4-H11-5 | 59.7 | 0.0768 | 5.89 | |||||||||||
| qGL12 | RM1337 | –RM3739 | 59.7 | 0.2160 | 11.04 | |||||||||||
| 粒宽 | qGW1 | H1-5 | –RM8097 | 141.6 | –0.0396 | 3.51 | ||||||||||
| Grain width | qGW2.1 | H2-6 | –H2-7 | 47.0 | 0.1134 | 9.58 | ||||||||||
| qGW2.2 | H2-7 | –RM262 | 52.7 | 0.0977 | 9.34 | |||||||||||
| qGW5 | RM17954 | –H5-7 | 27.5 | 23.5 | 0.1202 | 0.1143 | 12.28 | 11.44 | qGW5;JMJ703 | |||||||
| qGW6 | H6-2 | –RM3496 | 47.0 | 0.0728 | 7.06 | |||||||||||
| 粒厚 | qGT1 | RM8097 | –RM6703 | 163.5 | 163.5 | –0.1074 | –0.0907 | 12.75 | 12.59 | |||||||
| Grain thickness | qGT2.1 | H2-3 | –H2-4 | 33.2 | 0.0919 | 9.01 | ||||||||||
| qGT2.2 | H2-5 | –H2-6 | 41.8 | 0.0823 | 9.51 | |||||||||||
| qGT6 | wgwl | –RM3183 | 35.2 | 0.0708 | 8.21 | |||||||||||
| qGT9 | RM242 | –H9-6 | 69.3 | 0.0830 | 9.69 | |||||||||||
| 长宽比 | qGLW2.1 | H2-4 | –RM300 | 34.9 | –0.0787 | 13.88 | ||||||||||
| Length to width | qGLW2.2 | H2-5 | –H2-6 | 42.8 | –0.1332 | 15.61 | ||||||||||
| qGLW2 | H2-6 | –H2-7 | 50.0 | –0.0496 | 12.95 | |||||||||||
| qGLW5 | RM17954 | –H5-7 | 28.5 | 27.5 | –0.1598 | –0.1302 | 17.25 | 12.75 | ||||||||
| qGLW11 | RM167 | –H11-2 | 30.9 | 0.0519 | 7.79 | |||||||||||
| qGLW12 | RM1337 | –RM3739 | 51.7 | 0.0975 | 6.25 | |||||||||||
| 千粒重 | qTGW1.1 | H1-5 | –RM8097 | 144.6 | –1.1430 | 8.17 | OsTRBF3 | |||||||||
| 1000-grain | qTGW1.2 | RM8097 | –RM6703 | 148.5 | –1.0656 | 8.43 | ||||||||||
| weight | qTGW4 | RM317 | –RM5879 | 113.5 | 1.1454 | 7.41 | ||||||||||
| qTGW5 | RM18751 | –RM3476 | 85.1 | –1.2472 | 10.55 | |||||||||||
| qTGW9 | H9-4 | –RM3700 | 52.6 | –0.8719 | 4.55 | |||||||||||
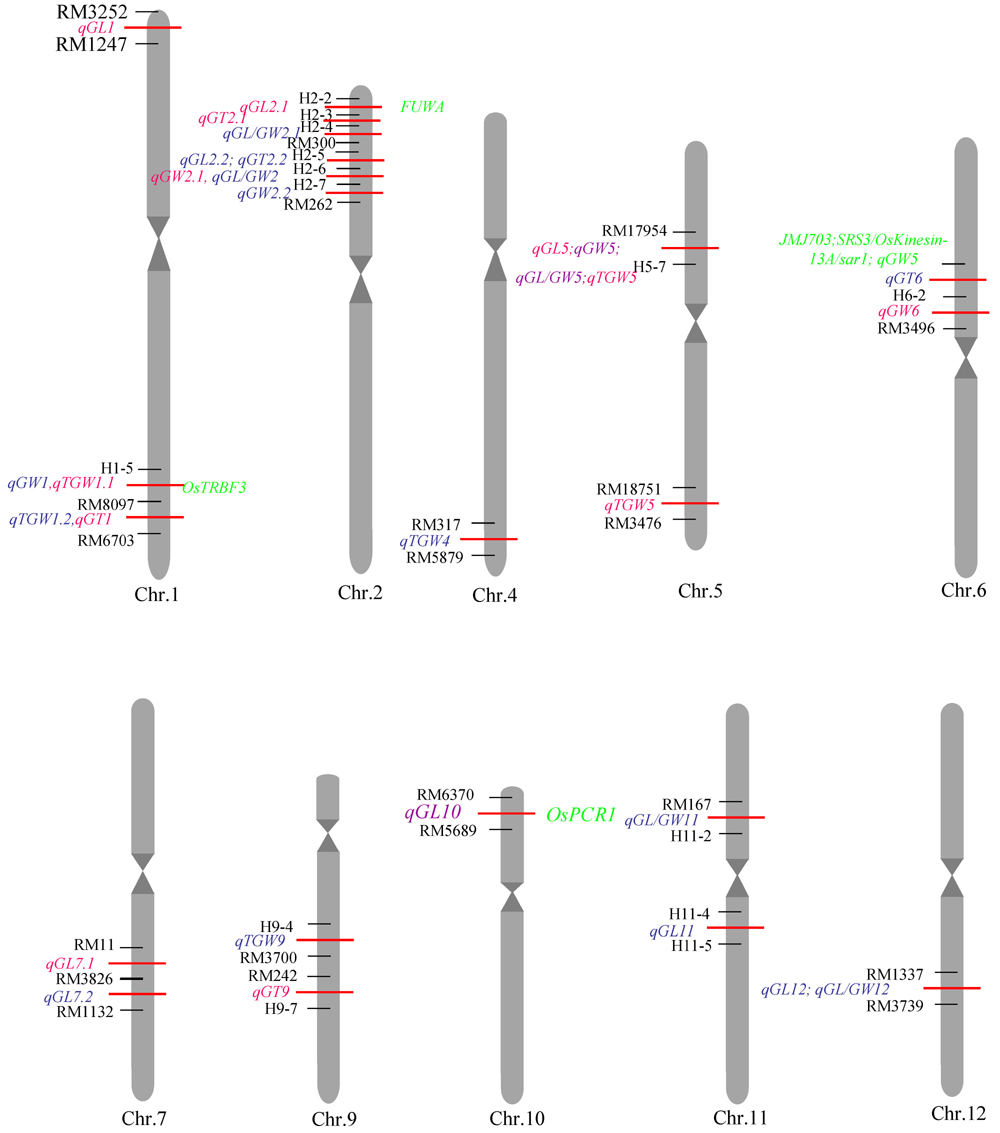
Fig.2. Distribution of QTLs related to grain shape traits in rice chromosome. Red labels indicated the QTLs detected in Hangzhou in 2016, and the blue labelsthe QTLs detected in Lingshui in 2016, the green labels indicated rice-related cloned genes in the interval.
| 基因名称基因登录号 Gene name Gene ID | 位置 Location | 春江16B CJ16B | 春江16B-AA CJ16B-AA | C84 | C84-AA | 变异类型 Variation type |
|---|---|---|---|---|---|---|
| FUWALOC_Os02g13950 | 9-11 | CCT | Pro | F | ||
| 120 | A | AAA/Lys | G | AAG/Lys | Si | |
| qSW5 LOC_Os05g09520 | 986 | C | CCG/Pro | T | CTG/Leu | S |
| 1014 | G | CGC/Arg | T | CTC/Leu | S | |
| 1072 | G | GCG/Ala | A | ACG/Thr | S | |
| 1100 | C | GCG/Ala | T | GTG/Val | S | |
| OsPCR1 LOC_Os10g02300 | 172 | G | GCC/Ala | T | TCC/Ser | S |
| 467 | A | CAC/His | T | CTC/Leu | S | |
| 507-509 | GAG | F | ||||
| 559-561 | TAA | STOP | CAC | His | S | |
| SRS3LOC_Os05g06280 /Kinesin-13A /Sar1 | 1189 | C | CTG/Leu | T | TTG/Leu | Si |
| 2420 | G | CGT/Arg | A | CAT/His | S | |
| 2505 | A | GAA/Glu | C | GAC/Asp | S | |
| OsTRBF3 LOC_Os01g51154 | 542 | G | TGA/STOP | T | TTA/Leu | S |
| 547-548 | CC | CCA/Pro | AG | AGA/Arg | S | |
| 554 | T | TTT/Phe | A | TAT/Tyr | S | |
| 556-557 | TG | TGG/Trp | GT | GTG/Val | S | |
| 561 | T | CAT/His | G | CAG/Gln | S | |
| 565 | G | GGG/Gly | A | AGG/Arg | S |
Table 5 Sequence variance analysis of the cloned grain shape main effect QTL allele in the two parents (C84 and CJ16B).
| 基因名称基因登录号 Gene name Gene ID | 位置 Location | 春江16B CJ16B | 春江16B-AA CJ16B-AA | C84 | C84-AA | 变异类型 Variation type |
|---|---|---|---|---|---|---|
| FUWALOC_Os02g13950 | 9-11 | CCT | Pro | F | ||
| 120 | A | AAA/Lys | G | AAG/Lys | Si | |
| qSW5 LOC_Os05g09520 | 986 | C | CCG/Pro | T | CTG/Leu | S |
| 1014 | G | CGC/Arg | T | CTC/Leu | S | |
| 1072 | G | GCG/Ala | A | ACG/Thr | S | |
| 1100 | C | GCG/Ala | T | GTG/Val | S | |
| OsPCR1 LOC_Os10g02300 | 172 | G | GCC/Ala | T | TCC/Ser | S |
| 467 | A | CAC/His | T | CTC/Leu | S | |
| 507-509 | GAG | F | ||||
| 559-561 | TAA | STOP | CAC | His | S | |
| SRS3LOC_Os05g06280 /Kinesin-13A /Sar1 | 1189 | C | CTG/Leu | T | TTG/Leu | Si |
| 2420 | G | CGT/Arg | A | CAT/His | S | |
| 2505 | A | GAA/Glu | C | GAC/Asp | S | |
| OsTRBF3 LOC_Os01g51154 | 542 | G | TGA/STOP | T | TTA/Leu | S |
| 547-548 | CC | CCA/Pro | AG | AGA/Arg | S | |
| 554 | T | TTT/Phe | A | TAT/Tyr | S | |
| 556-557 | TG | TGG/Trp | GT | GTG/Val | S | |
| 561 | T | CAT/His | G | CAG/Gln | S | |
| 565 | G | GGG/Gly | A | AGG/Arg | S |
| 基因名称 Gene name | 位置 Location | 限制性内切酶Restriction enzyme | 酶切位点Enzyme cutting site | 引物Primer |
|---|---|---|---|---|
| FUWA | 120 | SalⅠ | GTCGAC | F: ACGACGACGACGACGACGAAGGAACCCGCGTCGA |
| R: CCTCCTCCTCCTCCTCCTC | ||||
| OsPCR1 | 467 | StuⅠ | AGGCCT | F: TCGCACAGATGCACCGTGAGCTCAAGAACCGAGGCC |
| R: GTGTTGGGGAGGAGTCATCA | ||||
| OsTRBF3 | 565 | MaeⅡ | ACGT | F: ATCCTTCTTTGGACTTGGAGACTTATTTGCTTACG |
| R: TGCTTCTAGGGCAGGACTCG | ||||
| qGW5 | 986 | HaeⅢ | GGCC | F: CGAGGACTGGTGCGCCAACTCCATGTCGTGGC |
| R: GTTGGACATGTAGTTCGGGC | ||||
| SRS3/OsKinesin-13A/sar1 | 1189 | HindⅢ | AAGCTT | F: CGCCGCGCAGCTCGAGCTCTAAAGCT |
| R: GGTTTCAGCATCGCCTCAAA |
Table 6 dCAPsmolecular marker designed according totarget gene differencesite of the two parents C84 and CJ16B.
| 基因名称 Gene name | 位置 Location | 限制性内切酶Restriction enzyme | 酶切位点Enzyme cutting site | 引物Primer |
|---|---|---|---|---|
| FUWA | 120 | SalⅠ | GTCGAC | F: ACGACGACGACGACGACGAAGGAACCCGCGTCGA |
| R: CCTCCTCCTCCTCCTCCTC | ||||
| OsPCR1 | 467 | StuⅠ | AGGCCT | F: TCGCACAGATGCACCGTGAGCTCAAGAACCGAGGCC |
| R: GTGTTGGGGAGGAGTCATCA | ||||
| OsTRBF3 | 565 | MaeⅡ | ACGT | F: ATCCTTCTTTGGACTTGGAGACTTATTTGCTTACG |
| R: TGCTTCTAGGGCAGGACTCG | ||||
| qGW5 | 986 | HaeⅢ | GGCC | F: CGAGGACTGGTGCGCCAACTCCATGTCGTGGC |
| R: GTTGGACATGTAGTTCGGGC | ||||
| SRS3/OsKinesin-13A/sar1 | 1189 | HindⅢ | AAGCTT | F: CGCCGCGCAGCTCGAGCTCTAAAGCT |
| R: GGTTTCAGCATCGCCTCAAA |
| [1] | TianZ X,QianQ, LiuQ Q, YanM X, LiuX F, Yan C J, Liu G F, Gao Z Y, Tang S Z, Zeng D L, Wang Y H, Yu J M, Li J Y.Allelic diversities in rice starch biosynthesis lead to a diverse array of rice eating and cooking qualities.Proc NatlAcadSciUSA,2009, 106(51): 21760-21765. |
| [2] | 李孝琼, 韦宇, 高国庆, 邓国富, 郭嗣斌. 水稻遗传图谱构建及粒形相关性状的QTL定位. 南方农业学报, 2014, 45(7): 1154-1159. |
| Li X Q, Wei Y, Gao G Q, Deng G F, Guo C B.Construction of genetic map and mapping quantitative trait loci for grain shape-related traits in rice.JSouthAgric,2014, 45(7):1154-1159.(in Chinese with English abstract) | |
| [3] | McCouch S R,Teytelman L,Xu Y,Lobos K B, Clare K,Walton M, Fu B, Maghirang R, Li Z, Xing Y, Zhang Q, Kono L, Yano M, Fjellstrom R, DeClerck G, Schneider D, Cartinhour S, Ware D, Stein L.Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.) .DNA Res, 2002, 9(6):257-279. |
| [4] | Xu J C,Zhu L, Chen Y,LuC F, CaiH W.Construction of a rice molecular linkage map using a double hploid (DH) population.JGenGenom, 1994,21(3):205-214. |
| [5] | 张启军, 叶少平, 虞德容, 李平, 吕川根, 邹江石. 六张水稻遗传连锁图谱的比较分析. 西南农业学报,2005, 18(5): 584-592. |
| Zhang Q J, Ye S P, Yu D R, Li P, Lu C G, Zou J S.Comparison of six rice genetic linkage maps.Southwest China JAgricSci, 2005, 18(5): 584-592.(in Chinese with English abstract) | |
| [6] | 庄杰云, 施勇烽, 应杰政, 鄂志国, 曾瑞珍, 陈洁, 朱智伟. 中国主栽水稻品种微卫星标记数据库的初步构建. 中国水稻科学, 2006, 20(5):460-468. |
| Zhuang J Y, Shi Y F, Ying J Z, Dong Z G, Zeng R Z, Chen J, Zhu Z W.Construction and testing of primary microsatellite database of major rice varieties in China.Chin J Rice Sci, 2006, 20(5): 460-468.(in Chinese with English abstract) | |
| [7] | Xu J J, Zhao Q, Du P N, Xu C W,LiuQ Q, FengQ, WangB H, Tang S Z, Gu M H, Han B, Liang G H. Developing high throughput genotyped chromosome segment substitution lines based on population whole-genome re-sequencing in rice (Oryza sativa L.).BMC Genomics, 2010,11(1): 656. |
| [8] | 沈利爽, 何平, 徐云碧, 谭震波, 陆朝福, 朱立煌.水稻DH群体的分子连锁图谱及基因组分析. 植物生态学报,1998, 40(12):1115-1122. |
| Shen L S, He P, Xu Y B, Tan Z B, Lu C F, Zhu L H.Genetic molecular linkage map construction and genome analysis of rice doubled.Plant ActaEcol Sin, 1998, 40(12):1115-1122. (in Chinese with English abstract) | |
| [9] | Huang R Y, Jiang L G, Zheng J S, Wang T S, Huang Y M, Wang H C, Hong Z L.Genetic bases of rice grain shape: So many genes, so little known.TrendsPlant Sci, 2013, 18(4):218-226. |
| [10] | 方先文,张云辉, 肖西林,张所兵,林静,汪迎节.基于重组自交系群体的水稻粒形QTL定位. 江苏农业学报, 2017, 33(2): 241-247. |
| Fang X W, Zhang Y H, Xiao X L, Zhang S B, Lin J, Wang Y J.Mapping of QTLs for grain shape using recombinant inbred lines in rice ( Oryza sativa L.).ActaAgric Jiangsu, 2017,33(2): 241-247.(in Chinese with English abstract) | |
| [11] | Xia D, Zhou H, Qiu L, Jiang H, Zhang Q L, Gao G J, He Y Q.Mapping and verification of grain shape QTLs based on an advanced backcross population in rice.PloS One, 2017, 12(11):e0187553. |
| [12] | Zhao D, Li P B, Wang L Q, Sun L, Xia D, Luo L J, Gao G J, He Y Q, Zhang Q L.Genetic dissection of large grain shape in rice cultivar ‘Nanyangzhan’ and validation of a grain thickness QTL (qGT3.1) and a grain length QTL(qGL3.4).MolBreed, 2017, 37(3):42. |
| [13] | Zhou L Q,Wang Y P, Li S G.Genetic analysis and physical mapping of Lk-4 (t), a major gene controlling grain length in rice, with a BC2F2 population.Acta GenSin, 2006,33(1):72-79. |
| [14] | Wang Y X, Xiong G S, Hu J, JiangL, YuH, Fang Y X, Xu E B, Ye W J, Liu R F, Jing Y H, Zhu X D, Qian Q, Jing Y H, Xu J, Zeng L J, Xu J, Meng X B, Chen H Q, Wang Y H, Li J Y. Copy number variation at the GL7 locus contributes to grain size diversity in rice.NatGenet, 2015, 47(8):944-948. |
| [15] | Wu T, Shen Y Y, Zheng M, ChenY L, FengZ M, Liu X, Chen Z J, Wang J L, Wan J M, Yang C Y, Liu S J, Lei C L, Jiang L. Gene SGL SGL.Plant Cell Rep, 2014, 33(2):235-244. |
| [16] | Takano-Kai N, Doi K, Yoshimura A. GS3# participates in stigma exsertion as well as seed length in rice. BreedSci#, 2011, 61(3):244-250. |
| [17] | Qi P, Lin Y S, Song X J, ShenJB, HuangW, Shan J X, Zhu M Z, Jiang L W, Gao J P, Lin H X. The novel quantitative trait locus GL3.1 controls rice grain size and yield by regulating Cyclin-T1;3.Cell Res, 2012, 22(12):1666-1680. |
| [18] | Wu W,Liu X, Wang M,MeyerRS, LuoX, NdjiondjopMN, Tan L, Zhang J, Wu J, Cai H, Sun C, Wang X, Wing R A, Zhu Z.A single-nucleotide polymorphism causes smaller grain size and loss of seed shattering during African rice domestication.NatPlants, 2017, 3:17064. |
| [19] | Li Y B, Fan C C, Xing Y Z, JiangY H, LuoL J,SunL, Shao D, Xu C J, Li X H, Xiao J X, He Y Q, Zhang Q F.Natural variation in GS5 plays an important role in regulating grain size and yield in rice.NatGenet,2011, 43(12):1266-1292. |
| [20] | Song X J, Huang W, Shi M, ZhuMZ, LinHX.A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase.NatGenet, 2007, 39(5):623-630. |
| [21] | Yan S, Zou G H, Li S J, Wang H, LiuH Q, ZhaiG W, Guo P, Song H M, Yan C J, Tao Y Z. Seed size is determined by the combinations of the genes controlling different seed characteristics in rice.TheorApplGenet, 2011, 123(7):1173-1181. |
| [22] | Liu J F, Chen J, Zheng X M, Wu F Q, Lin Q B, Heng Y Q, Tian Peng, Cheng Z J, Yu X W, Zhou K N, Zhang X, Guo X P, Wang J L, Wang H Y, Wan J M.GW5 acts in the brassinosteroidsignalling pathway to regulate grain width and weight in rice. NatPlants, 2017, 3:17043. |
| [23] | Xue W Y, Xing Y Z, Weng X Y, Zhao Y, Tang W J, Zhou H J, Yu S B, Xu C B, Li X H, Zhang Q F.Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice.NatGenet, 2009, 40(6):761-767. |
| [24] | LiJ,ThomsonM, McCouchSR. Fine mapping of a grain-weight quantitative trait locus in the pericentromeric region of rice chromosome 3.Genetics, 2004,168(4): 2187. |
| [25] | Bai X F, Luo L J, Yan W H,Mallikarjuna R K, Zhan W, Xing Y Z.Genetic dissection of rice grain shape using a recombinant inbred line population derived from two contrasting parents and fine mapping a pleiotropic quantitative trait locus qGL7.BMC Genet, 2010, 11(1): 16. |
| [26] | Shao G N, Tang S Q, Luo J, Jiao G A, Wei X J, Tang A, Wu J L, Zhuang J Y, Hu P S.Mapping of qGL7-2, a grain length QTL on chromosome 7 of rice.JGenGenom, 2010, 37(8):523-531. |
| [27] | Liu T M, Shao D, Kovi M R, Zhong X Y.Mapping and validation of quantitative trait loci for spikelets per panicle and 1,000-grain weight in rice ( Oryza sativa L.).TheorApplGenet, 2010, 120(5):933-942. |
| [28] | Xie X, Jin F, Song M H, Suh J P, Hwang H G.Fine mapping of a yield-enhancing QTL cluster associated with transgressive variation in an Oryza sativa ×O. rufipogoncross. TheorApplGenet, 2008, 116(5):613-622. |
| [29] | 陈建华, 姚青, 谢绍军,孙成效,朱智伟. 机器视觉在稻米粒型检测中的应用.中国水稻科学, 2007, 21(6):669-672. |
| Chen J H,Yao Q, Xie S J,Sun C X, Zhu Z W.Detection of rice shape based on machine vision.Chin J Rice Sci, 2007, 21(6):669-672. (in Chinese with English abstract) | |
| [30] | 刘仁虎, 孟金陵. MapDraw在Excel中绘制遗传连锁图的宏.遗传, 2003, 25(3):317-321. |
| Liu R H, Meng J L.MapDraw: Amicro for drawing genetic linkage maps based on given genetic linkage data.Hereditas, 2003, 25(3):317-321. (in Chinese with English abstract) | |
| [31] | McCouch S R. Genenomenclaturesystem forrice.Rice,2008, 1(1):72-84. |
| [32] | Song W Y, Lee H S, Jin S R, KoD, Martinoia E, LeeY. RicePCR1 influences grain weight and Zn accumulation in grains. Plant Cell & Environ,2015, 38(11):2327-2339. |
| [33] | Chen J, Gao H, Zheng X M, JinM, WengJF, MaJ. An evolutionarily conserved gene, FUWA, plays a role in determining panicle architecture, grain shape and grain weight in rice.Plant J, 2015, 83(3):427-438. |
| [34] | Liu X Y, Zhou S L, Wang W T, YeY R, ZhaoY, XuQ T, Zhou C, Tan F, Cheng S F, Zhou D X.Regulation ofhistone methylation and reprogramming of gene expression in the rice inflorescence meristem. Plant Cell, 2015, 27(5):1428-1444. |
| [35] | ZhuYD,LingTL,TangL, SongY, BaiJK, ShanJH, Chang J Y, Tai W.OsKinesin-13A is an active microtubule depolymerase involved in glume length regulation via affecting cell elongation.SciRep, 2015, 5(10):9457. |
| [36] | Byun M Y, Hong J P, KimW T. Identification and characterization of three telomere repeat-binding factors in rice.BiochemBiophyResComm, 2008, 372(1):85-90. |
| [37] | Cui X K, Jin P, Cui X,Gu L F, Lu Z K, Xue Y M, Wei L Y, Qi J F, Song X W, Luo M, An G, Cao X F.Control of transposon activity by a histone H3K4 demethylase in rice.Proc NatlAcadSci USA, 2013, 110(5):1953-1958. |
| [38] | Kitagawa K, Kurinami S, Oki K, Abe Y, Ando T, Kono I, Yano M, Kitano H, Iwasaki Y.A novel Kinesin 13 protein regulating rice seed length.Plant & Cell Physiol, 2010, 51(8):1315-1329. |
| [39] | Yoshida S, Ikegami M, Kuze J, Sawada K, Nashimoto Z, Nakamura C, Ishii T, Kamijima O.QTL analysis for plant and grain characters of sake-brewing rice using a doubled haploid population.Breed Sci, 2002, 52(4):309-317. |
| [40] | Li Z F, Wan J M, Xia J F,Zhai H Q.Mapping quantitative trait loci underlying appearance quality of rice grains (Oryza sativa L.). JGenGenom,2003, 30(3):251-259. |
| [41] | Huang N, Parco A, Mew T,Magpantay G, McCouch S, Guiderdoni E, Xu J, Subudhi P, Angeles E R, Khush G S. RFLP mapping of isozymes, RAPD and QTLs for grain shape, brown planthopper resistance in a doubled haploid rice population.MolBreed, 1997, 3(2):105-113. |
| [42] | Wan X Y, Wan J M, Weng J F,Jiang L, Bi J C, Wang C M, Zhai H Q.Stability of QTLs for rice grain dimension and endosperm chalkiness characteristics across eight environments.TheorApplGenet, 2005, 110(7):1334. |
| [43] | Hittalmani S, Huang N, Courtois B,Venuprassad R, Shashidhar H E, Zhuang J Y, Zheng K L, Liu G F, Wang G C, Sidhu J S, Srivantaneeyakul S, Sinqh V P, Baqali P G, Prasanna H C, McLaren G, Khush G S. Identification of QTL for growth- and grain yield-related traits in rice across nine locations of Asia.TheorApplGenet, 2003, 107(4):679-690. |
| [44] | GaoYM, ZhuJ, SongYS, He C X, Shi C H, Xing Y Z. Analysis of digenic epistatic effects and QE interaction effects QTL controlling grain weight in rice.JZhejiang UnivSci, 2004, 5(4):371-377. |
| [45] | Lin H X, Qian H R, Zhuang J Y, Lu J, Min S K, Xiong Z M, Huang N, Zheng K L.RFLP mapping of QTLs for yield and related characters in rice ( Oryza sativa L.).TheorApplGenet, 1996, 92(8):920-927. |
| [46] | ChoYC, SuhJP, ChoiIS, Hong H C, Baek M K, Hong H C, Kim Y G, Choi H, Moon H P, Hwang G H. QTLs analysis of yield and its related traits in wild rice relative Oryzarufipogon.TreatCropRes, 2003(4): 19-29. |
| [47] | Hua J, Xing Y, Wu W, Xu C, Sun X, Yu S, Zhang Q.Single-locus heterotic effects and dominance by dominance interactions can adequately explain the genetic basis of heterosis in an elite rice hybrid.Proc NatlAcadSciUSA, 2003, 100(5):2574. |
| [48] | Zhuang J Y, Fan Y Y, Rao Z M, Wu J L, Xia Y W, Zheng K L.Analysis on additive effects and additive-by-additive epistatic effects of QTLs for yield traits in a recombinant inbred line population of rice.TheorApplGenet, 2002, 105(8):1137-1145. |
| [49] | Hua J P, Xing Y Z, Xu C G, Sun X L, Yu S B, Zhang Q.Genetic dissection of an elite rice hybrid revealed that heterozygotes are not always advantageous for performance.Genetics, 2003, 162(4):1885-1895. |
| [50] | 谭震波, 沈利爽, 陆朝福, 陈英, 朱立煌, 周开达, 袁祚廉. 水稻再生能力和头季稻产量性状的QTL定位及其遗传效应分析. 作物学报, 1997, 23(3): 289-295. |
| Tan Z B,Shen L S, Yuan Z L, Lu C F, Chen Y, Zhou K D, Zhu L H, Zuo W X B. Identification of QTLs for ratooning ability and grain yield traits of rice and analysis of their genetic effects.ActaAgronSin, 1997, 23(3):289-295. (in Chinese with English abstract) |
| [1] | Zhibin CAO, Yao LI, Bohong ZENG, Linghua MAO, Yaohui CAI, Xiaofeng WU, Linfeng YUAN. QTL Mapping for Heat Tolerance of Chalky Grain Rate of Oryza glaberrima Steud. [J]. Chinese Journal OF Rice Science, 2020, 34(2): 135-142. |
| [2] | Xiaolei WANG, Yang LIU, Xiaotang SUN, Linjuan OUYANG, Jinlong PAN, Xiaosong PENG, Xiaorong CHEN, Xiaopeng HE, Junru FU, Jianmin BIAN, Lifang HU, Jie XU, Haohua HE, Changlan ZHU. Identification and Stability Analysis of QTL for Grain Quality Traits Under Multiple Environments in Rice [J]. Chinese Journal OF Rice Science, 2020, 34(1): 17-27. |
| [3] | Andong ZHU, Zhichao SUN, Yujun ZHU, Hui ZHANG, Xiaojun NIU, Yeyang FAN, Zhenhua ZHANG, Jieyun ZHUANG. Identification of QTL for Grain Weight and Grain Shape Using Populations Derived from Residual Heterozygous Lines of indica Rice [J]. Chinese Journal OF Rice Science, 2019, 33(2): 144-151. |
| [4] | Jin LIU, Xiaoyun YAO, Dan LIU, Liqin YU, Hui LI, Qi WANG, Jiayu WANG, Maomao LI. Identification of QTL for Panicle Traits under Multiple Environments in Rice(Oryza sativa L.) [J]. Chinese Journal OF Rice Science, 2019, 33(2): 124-134. |
| [5] | Biaolin HU, Derun HUANG, Yeqing XIAO, Qiangsheng HE, Yong WAN, Yeyang FAN. QTL Analysis for Mineral Contents in Brown Rice Using a BC2F4:5 Population Derived from Dongxiang Wild Rice (Oryza rufipogon Griff.) [J]. Chinese Journal OF Rice Science, 2018, 32(1): 43-50. |
| [6] | Han LIN, Jiangmin XU, Huqian HU, An ZHENG, Wanlu XU, Ping LOU, Yuexing WANG, Dali ZENG, Yuchun RAO. Identifying of QTLs for Resistance to Metal Irons Stress in Rice [J]. Chinese Journal OF Rice Science, 2018, 32(1): 23-34. |
| [7] | Guilian ZHANG, Bin LIAO, Wenbang TANG, Liyun CHEN, Yinghui XIAO. Identifying QTLs for Thermo-tolerance of Grain Chalkiness Trait in Rice [J]. Chinese Journal OF Rice Science, 2017, 31(3): 257-264. |
| [8] | Jun WANG, Jin-yan ZHU, Yong ZHOU, Jie YANG, Fang-jun FAN, Wen-qi LI, Fang-quan WANG, Wei-gong ZHONG, Guo-hua LIANG. QTL Analysis for Heading Date in Rice (Oryza sativa L.) Under Different Temperatures and Light Intensities [J]. Chinese Journal OF Rice Science, 2016, 30(3): 247-255. |
| [9] | Chang-feng ZHU, Li-jun LIANG, Si-yuan ZENG, Tian-wei LI, Guan-shan DONG, De-lin HONG. Fine Mapping of qFla-8-2 for Flag Leaf Angle in Rice [J]. Chinese Journal OF Rice Science, 2016, 30(1): 27-34. |
| [10] | Zhi-bin CAO, Hong-wei XIE, Yuan-yuan NIE, Ling-hua MAO, Yong-hui LI, Yao-hui CAI. Mapping a QTL(qHTH5) for Heat Tolerance at the Heading Stage on Rice Chromosome 5 and Its Genetic Effect Analysis [J]. Chinese Journal OF Rice Science, 2015, 29(2): 119-125. |
| [11] | Jie REN, Xiu-qin ZHAO, Zai-song DING, Chao XIANG, Jing ZHANG, Chao WANG, Jun-wei ZHANG, Augustino JOSEPH Charles, Qiang ZHANG, Yun-long PANG, Yong-ming GAO, Ying-yao SHI. Dissection and QTL Mapping of Low-phosphorus Tolerance Using Selected Introgression Lines in Rice [J]. Chinese Journal OF Rice Science, 2015, 29(1): 1-13. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||