
Chinese Journal OF Rice Science ›› 2018, Vol. 32 ›› Issue (2): 103-110.DOI: 10.16819/j.1001-7216.2018.7056
• Research Papers • Next Articles
Xixu PENG1,2, Ningning BAI1, Haihua WANG1,2,*
Received:2017-05-20
Online:2018-03-10
Published:2018-03-10
Contact:
Haihua WANG
彭喜旭1,2, 白宁宁1, 王海华1,2,*
通讯作者:
王海华
基金资助:CLC Number:
Xixu PENG, Ningning BAI, Haihua WANG. Isolation and Expression Profiles of Cadmium Stress-Responsive Rice WRKY15 Transcription Factor Gene[J]. Chinese Journal OF Rice Science, 2018, 32(2): 103-110.
彭喜旭, 白宁宁, 王海华. 响应镉胁迫的水稻WRKY15转录因子基因的分离与表达特征[J]. 中国水稻科学, 2018, 32(2): 103-110.
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.ricesci.cn/EN/10.16819/j.1001-7216.2018.7056
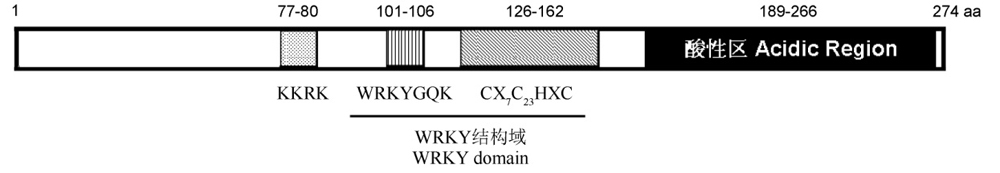
Fig. 1. Schematic diagram of the deduced protein of the OsWRKY15 gene. Dotted, vertically lined, diagonally lined and filled bars represent nuclear localization signal, WRKYGQK peptide, zinc-finger motif and acidic region, respectively. Conserved WRKY domain and zinc-finger pattern are presented, and numbers indicate the amino acid position of each structure in OsWRKY15.
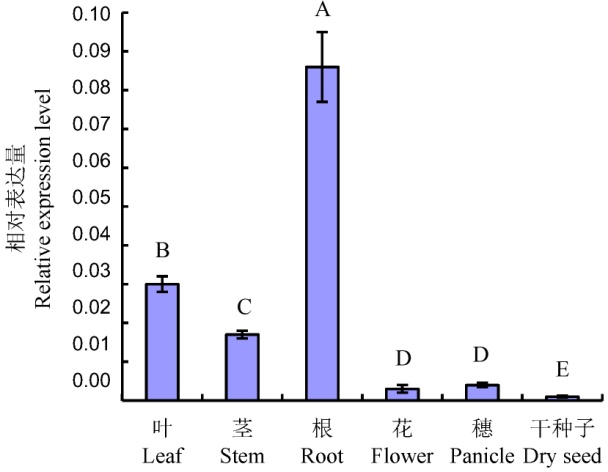
Fig. 3. Expression patterns of OsWRKY15 in different rice organs. Different uppercase letters indicate significant difference at 0.01 level (n=6) according to Duncan’s multiple range test.
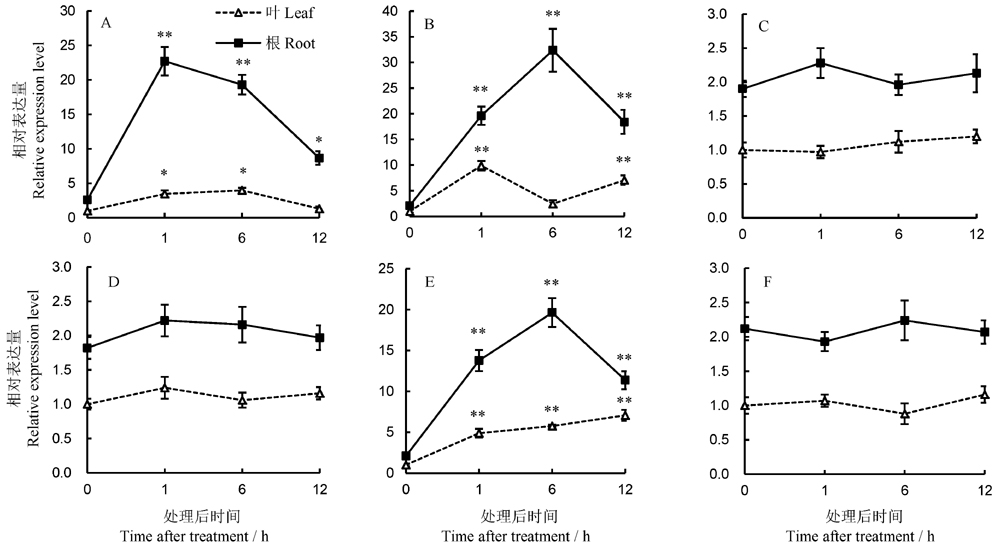
Fig. 4. OsWRKY15 expression in response to different treatments with hormones and Cd exposure. A, Cadmium; B, Nitric oxide; C, Salicylic acid; D, Methyl jasmonate; E, Abscisic acid; F, Ethephon; *, ** indicate significant difference at 0.05, 0.01 level(n=6), respectively, in relative to that of the control at 0 h according to Duncan’s multiple range test.
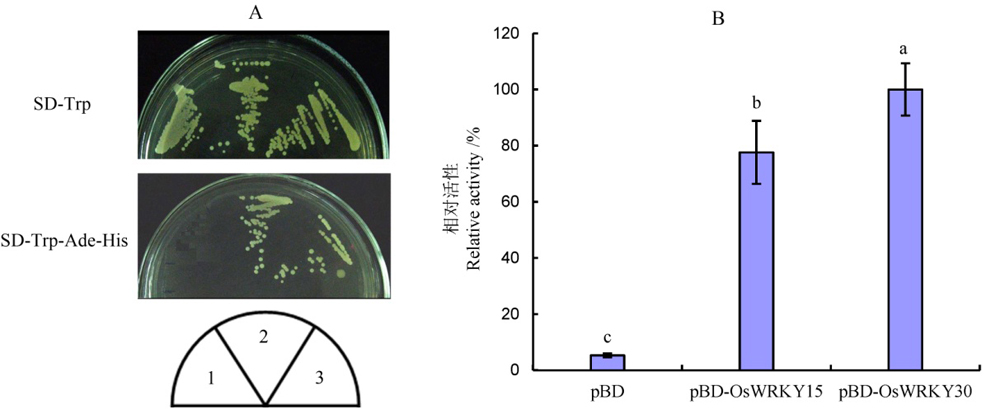
Fig. 5. Assay of transcriptional activation activity of OsWRKY15 in yeast cells. A, Yeast transformants grew in SD-Trp or SD-Trp-Ade-His media. 1, Empty vector pGBKT7 (negative control); 2, pGBKT7-OsWRKY15; 3, pGBKT7-30 (positive control); B, Assay for relative β-galactosidase activity of yeast transformants. Different lowercase letters indicate significant difference at P<0.05 according to Duncan’s multiple range test.
| [1] | Cosio C, Vollenweider P, Keller C.Localization and effects of cadmium in leaves of a cadmium-tolerant willow (Salix viminalis L.): I. Macrolocalization and phytotoxic effects of cadmium. Environ Exp Bot, 2006, 58: 64-74. |
| [2] | Grant C A, Clarke J M, Duguid S, Chaney R L.Selection and breeding of plant cultivars to minimize cadmium accumulation.Sci Total Environ, 2008, 390: 301-310. |
| [3] | Folgar S, Torres E, Pérez-Rama M, Cid A, Herrero C, Abalde J.Dunaliella salina as marine microalga highly tolerant to but a poor remover of cadmium. J Hazard Mater, 2009, 165: 486-493. |
| [4] | Verbruggen N, Hermans C, Schat H.Mechanisms to cope with arsenic or cadmium excess in plants.Curr Opin Plant Biol, 2009, 12: 364-372. |
| [5] | Tang M F, Mao D H, Xu L W, Li D, Song S, Chen C.Integrated analysis of miRNA and mRNA expression profiles in response to Cd exposure in rice seedlings.BMC Genom, 2014, 15: 835. |
| [6] | Oono Y, Yazawa T, Kawahara Y, Kanamori H, Kobayashi F, Sasaki H, Mori S, Wu J, Handa H, Itoh T, Matsumoto T.Genome-wide transcriptome analysis reveals that cadmium stress signaling controls the expression of genes in drought stress signal pathways in rice.PLoS One, 2014, 9(5): e96946. |
| [7] | Farinati S, DalCorso G, Varotto S, Furini A. TheBrassica juncea BjCdR15, an ortholog of Arabidopsis TGA3, is a regulator of cadmium uptake, transport and accumulation in shoots and confers cadmium tolerance in transgenic plants. New Phytol, 2010, 185: 964-978. |
| [8] | van de Mortel J E, Schat H, Moerland P D, Ver Loren van Themaat E, van der Ent S, Blankestijn H, Ghandilyan A, Tsiatsiani S, Aarts M G. Expression differences for genes involved in lignin, glutathione and sulphate metabolism in response to cadmium in Arabidopsis thaliana and the related Zn/Cd-hyperaccumulator Thlaspi caerulescens. Plant Cell Environ, 2008, 31: 301-324. |
| [9] | Eulgem T, Rushton P J, Robatzek S, Somssich I E.The WRKY superfamily of plant transcription factors.Trends Plant Sci, 2000, 5, 199-206. |
| [10] | Chen L, Song Y, Li S, Zhang L, Zou C, Yu D.The role of WRKY transcription factors in plant abiotic stresses.Biochim Biophys Acta, 2012, 819(2): 120-128. |
| [11] | Ricachenevsky F K, Sperotto R A, Menguer P K, Fett J P.Identification of Fe-excess-induced genes in rice shoots reveals a WRKY transcription factor responsive to Fe, drought and senescence.Mol Biol Rep, 2010, 37: 3735-3745. |
| [12] | Castrillo G, Sánchez-Bermejo E, de Lorenzo L, Crevillén P, Fraile-Escanciano A, Tc M, Mouriz A, Catarecha P, Sobrino-Plata J, Olsson S, Leo Del Puerto Y, Mateos I, Rojo E, Hernández L E, Jarillo J A, Piñeiro M, Paz-Ares J, Leyva A. WRKY6 transcription factor restricts arsenate uptake and transposon activation inArabidopsis. Plant Cell, 2013, 25: 2944-2957. |
| [13] | Ding Z J, Yan J Y, Xu X Y, Li G X, Zheng S J.WRKY46 functions as a transcriptional repressor of ALMT1, regulating aluminum-induced malate secretion in Arabidopsis. Plant J, 2013, 76: 825-835. |
| [14] | Jiang Y J, Yu D Q.WRKY transcription factors: Links between phytohormones and plant processes.Sci China Life Sci, 2015, 58: 501-502. |
| [15] | Lamotte O, Courtois C, Barnavon L, Pugin A, Wendehenne D.Nitric oxide in plants: The biosynthesis and cell signalling properties of a fascinating molecule.Planta, 2005, 221: 1-4. |
| [16] | Xiong J, Fu G, Tao L, Zhu C.Roles of nitric oxide in alleviating heavy metal toxicity in plants.Arch Biochem Biophys, 2010, 497: 13-20. |
| [17] | Hong C Y, Cheng D, Zhang G Q, Zhu D, Chen Y, Tan M.The role of ZmWRKY4 in regulating maize antioxidant defense under cadmium stress.Biochem Biophys Res Commun, 2017, 482: 1504-1510. |
| [18] | Wu K L, Guo Z J, Wang H H, Li J.The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Res, 2005, 12: 9-26. |
| [19] | 孟姣, 王海华, 向建华, 蒋丹, 彭喜旭, 贺欢欢. 水稻WRKY转录因子基因家族响应外源一氧化氮的表达谱分析. 中国水稻科学, 2016, 30(2): 111-120. |
| Meng J, Wang H H, Xiang J H, Jiang D, Peng X X, He H H.Expression profiles of rice WRKY transcription factor gene family responsive to exogenous nitric oxide application.Chin J Rice Sci, 2016, 30(2): 111-120. (in Chinese with English abstract) | |
| [20] | Peng X X, Hu Y, Tang X, Zhou P, Deng X, Wang H H, Guo Z.Constitutive expression of rice WRKY30 gene increases the endogenous jasmonic acid accumulation, PR gene expression and resistance to fungal pathogens in rice. Planta, 2012, 236: 1485-1498. |
| [21] | Schwechheimer C, Bevan M.The regulation of transcription factor activity in plants.Trends Plant Sci, 1998, 3: 378-383. |
| [22] | Asgher M, Khan M I, Anjum N A, Khan N A.Minimising toxicity of cadmium in plants-role of plant growth regulators.Protoplasma, 2015, 252: 399-413. |
| [23] | Wei W, Zhang Y, Han L, Guan Z, Chai T.A novel WRKY transcriptional factor from Thlaspi caerulescens negatively regulates the osmotic stress tolerance of transgenic tobacco. Plant Cell Rep, 2008, 27: 795-803. |
| [24] | Yang G, Wang C, Wang Y, Guo Y, Zhao Y, Yang C, Gao C.Overexpression of ThVHAc1 and its potential upstream regulator, ThWRKY7, improved plant tolerance of cadmium stress. Sci Rep, 2016, 6: 18752. |
| [25] | Li L, Wang Y, Shen W.Roles of hydrogen sulfide and nitric oxide in the alleviation of cadmium-induced oxidative damage in alfalfa seedling roots.Biometals, 2012, 25: 617e631. |
| [26] | Uraguchi S, Mori S, Kuramata M, Kawasaki A, Arao T, Ishikawa S.Root-to-shoot Cd translocation via the xylem is the major process determining shoot and grain cadmium accumulation in rice.J Exp Bot, 2009, 60: 2677-2688. |
| [27] | Ramamoorthy R, Jiang S Y, Kumar N, Venkatesh P N, Ramachandran S.A comprehensive transcriptional profiling of the WRKY gene family in rice under various abiotic and phytohormone treatments. Plant Cell Physiol, 2008, 49: 865-879. |
| [1] | HOU Xiaoqin, WANG Ying, YU Bei, FU Weimeng, FENG Baohua, SHEN Yichao, XIE Hangjun, WANG Huanran, XU Yongqiang, WU Zhihai, WANG Jianjun, TAO Longxing, FU Guanfu. Mechanisms Behind the Role of Potassium Fulvic Acid in Enhancing Salt Tolerance in Rice Seedlings [J]. Chinese Journal OF Rice Science, 2024, 38(4): 409-421. |
| [2] | TANG Zhiwei, ZHU Xiangcheng, ZHANG Jun, DENG Aixing, ZHANG Weijian. Effects of Green Manure Planting and Lime Application on Cadmium Content in Double-cropping Rice Under Controlled Irrigation [J]. Chinese Journal OF Rice Science, 2024, 38(2): 211-222. |
| [3] | HUANG Qina, XU Youxiang, LIN Guanghao, DANG Hongyang, ZHENG Zhenquan, ZHANG Yan, WANG Han, SHAO Guosheng, YIN Xianyuan. Effects of Silicon on Antioxidant Enzyme System and Expression Levels of Genes Related to Cd2+ Uptake and Transportation in Rice Seedlings Under Cadmium Stress [J]. Chinese Journal OF Rice Science, 2023, 37(5): 486-496. |
| [4] | CHENG Ling, HUANG Fugang, QIU Yipu, WANG Xinyi, SHU Wan, QIU Yongfu, LI Fahuo. Genetic Analysis and Identification of Brown Planthopper Resistance Gene in indica Rice Accession 570011 [J]. Chinese Journal OF Rice Science, 2023, 37(3): 244-252. |
| [5] | PEI Feng, WANG Guangda, GAO Peng, FENG Zhiming, HU Keming, CHEN Zongxiang, CHEN Hongqi, CUI Ao, ZUO Shimin. Evaluation of New japonica Rice Lines with Low Cadmium Accumulation and Good Quality Generated by Knocking Out OsNramp5 [J]. Chinese Journal OF Rice Science, 2023, 37(1): 16-28. |
| [6] | YUAN Yang, AO Hejun, ZHOU Zhonghua, YING Jiezheng, ZHANG Jian, NI Shen. Fine-mapping of SegD8 Loci for Rice Hybrid Segregation Distortion [J]. Chinese Journal OF Rice Science, 2023, 37(1): 37-44. |
| [7] | LI Xiaoxiu, LÜ Qiming, YUAN Dingyang. Research Progress on the Effects of OsNramp5 Mutation on Important Agronomic Traits in Rice [J]. Chinese Journal OF Rice Science, 2022, 36(6): 562-571. |
| [8] | SUN Zhiguang, DAI Huimin, CHEN Tingmu, LI Jingfang, CHI Ming, ZHOU Zhenling, LIU Yan, LIU Jinbo, XU Bo, XING Yungao, YANG Bo, LI Jian, LU Baiguan, FANG Zhaowei, WANG Baoxiang, XU Dayong. Phenotypic Identification and Gene Mapping of a Lesion Mimic Mutant lmm7 in Rice [J]. Chinese Journal OF Rice Science, 2022, 36(4): 357-366. |
| [9] | YAO Shu, ZHANG Yadong, LU Kai, WANG Cailin. Progress in Functions, Allelic Variations and Interactions of Soluble Starch Synthases Genes SSⅡa and SSⅢa in Rice [J]. Chinese Journal OF Rice Science, 2022, 36(3): 227-236. |
| [10] | LIANG Cheng, XIANG Xunchao, ZHANG Ouling, YOU Hui, XU Liang, CHEN Yongjun. Analyses on Agronomic Traits and Genetic Characteristics of Two New Plant-architecture Lines in Rice [J]. Chinese Journal OF Rice Science, 2022, 36(2): 171-180. |
| [11] | DONG Zheng, WANG Yamei, LI Yongchao, XIONG Haibo, XUE Canhui, PAN Xiaowu, LIU Wenqiang, WEI Xiucai, LI Xiaoxiang. Genome-wide Association Analysis of Cadmium Content in Rice Based on MAGIC Population [J]. Chinese Journal OF Rice Science, 2022, 36(1): 35-42. |
| [12] | Xiaobo XU, Penghu AN, Tianjiao GUO, Dan HAN, Wei JIA, Wuxing HUANG. Research Progresses on Response Mechanisms and Control Measures of Cadmium Stress in Rice [J]. Chinese Journal OF Rice Science, 2021, 35(5): 415-426. |
| [13] | Yuxiang LI, Hairong LIN, Qian LIANG, Guodong WANG. Effects of Dopamine Priming on Seed Germination and Seedling Growth of Rice Under Salt Stress [J]. Chinese Journal OF Rice Science, 2021, 35(5): 487-494. |
| [14] | Nan JIANG, Xu YAN, Yanbiao ZHOU, Qunfeng ZHOU, Kai WANG, Yuanzhu YANG. Factors Affecting Cadmium Accumulation in Rice and Strategies for Minimization [J]. Chinese Journal OF Rice Science, 2021, 35(4): 342-351. |
| [15] | Yujun ZHU, Ziwei ZUO, Zhenhua ZHANG, Yeyang FAN. A New Approach for Fine-mapping and Map-based Cloning of Minor-Effect QTL in Rice [J]. Chinese Journal OF Rice Science, 2021, 35(4): 407-414. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||