
中国水稻科学 ›› 2015, Vol. 29 ›› Issue (6): 578-586.DOI: 10.3969/j.issn.1001G7216.2015.06.003
袁筱萍, 王彩红, 邓宏中, 徐群, 冯跃, 余汉勇, 王一平, 魏兴华*( )
)
收稿日期:2015-01-19
修回日期:2015-09-17
出版日期:2015-10-25
发布日期:2015-11-10
通讯作者:
魏兴华
作者简介:*通讯录作者:E-mail:weixinghua@caas.cn
基金资助:
Xiao-ping YUAN, Cai-hong WANG, Hong-zhong DENG, Qun XU, Yue FENG, Han-yong YU, Yi-ping WANG, Xing-hua WEI*( )
)
Received:2015-01-19
Revised:2015-09-17
Online:2015-10-25
Published:2015-11-10
Contact:
Xing-hua WEI
About author:*Corresponding author:E-mail:weixinghua@caas.cn
摘要:
以69份类型明确的亚洲栽培稻品种为材料,选用120对SSR引物,通过评估遗传多样性和群体结构,研究分析其遗传变异对SSR引物数的要求。结果表明:1)120对引物在69份试验材料中共检测到1256个等位变异,每个位点的等位基因数差异较大,变化范围为2~27,平均为10.5;平均Nei基因多样性指数变化范围为0.4058~0.9452,平均为0.7616;2)分析亚洲栽培稻遗传多样性时,至少需要72对SSR引物;3)分析亚洲栽培稻群体结构时所需最少引物数可降低至60对。
中图分类号:
袁筱萍, 王彩红, 邓宏中, 徐群, 冯跃, 余汉勇, 王一平, 魏兴华. 亚洲栽培稻遗传变异分析最少SSR引物数的研究[J]. 中国水稻科学, 2015, 29(6): 578-586.
Xiao-ping YUAN, Cai-hong WANG, Hong-zhong DENG, Qun XU, Yue FENG, Han-yong YU, Yi-ping WANG, Xing-hua WEI. Minimum of SSR Markers for Analyzing Genetic Variation of Oryza sativa L.[J]. Chinese Journal OF Rice Science, 2015, 29(6): 578-586.
| 编号 Serial number | 品种名称 Accession name | 来源地 Country of origin | 类型 Classification | 编号 Serial number | 品种名称 Accession name | 来源地 Country of origin | 类型 Classification |
|---|---|---|---|---|---|---|---|
| ind-01 | Jaya | India | indica | aro-01 | JC101 | India | aromatic |
| ind-02 | IR8 | Phillipines | indica | aro-02 | Nga Ya Pauk | Myanmar | aromatic |
| ind-03 | Taichung Native 1 | Taiwan | indica | aro-03 | Kui Sali | Bangladesh | aromatic |
| ind-04 | Khao Dawk Mali 105 | Thailand | indica | aro-04 | DomSofid | Iran | aromatic |
| ind-05 | Padi Boenor | Indonesia | indica | aro-05 | Kalijira | Bangladesh | aromatic |
| ind-06 | JC92 | India | indica | aro-06 | Gordoi | Bangladesh | aromatic |
| ind-07 | Champa Tong 54 | Thailand | indica | aro-07 | JC73-4 | India | aromatic |
| ind-08 | Lageado | Brazil | indica | aro-08 | Shwewa | Myanmar | aromatic |
| ind-09 | Dhola Aman | Bangladesh | indica | aro-09 | Kanjari | India | aromatic |
| ind-10 | RTS12 | Vietnam | indica | aro-10 | Pankhari 203 | India | aromatic |
| ind-11 | IR36 | Phillipines | indica | aro-11 | N12 | India | aromatic |
| ind-12 | CO18 | India | indica | aro-12 | Basmati 1 | Pakistan | aromatic |
| ind-13 | Gie 57 | Vietnam | indica | trj-01 | Dou-Agu | Brazil | tropical japonica |
| ind-14 | Macan Binundok | Philippines | indica | trj-02 | Trembese | Indonesia | tropical japonica |
| ind-15 | TD25 | Thailand | indica | trj-03 | Gundil Kuning | Indonesia | tropical japonica |
| aus-01 | Shor Shori | Bangladesh | aus | trj-04 | Honduras | Brazil | tropical japonica |
| aus-02 | Jhona 349 | India | aus | trj-05 | Kinastano | Philippines | tropical japonica |
| aus-03 | Black Gora (Ncs12) | India | aus | trj-06 | Dholi Boro | Bangladesh | tropical japonica |
| aus-04 | Shada Boro | Bangladesh | aus | trj-07 | Cuba 65 | Cuba | tropical japonica |
| aus-05 | Unnamed | Bangladesh | aus | trj-08 | IAC25 | Brazil | tropical japonica |
| aus-06 | Badal 89 | Bangladesh | aus | trj-09 | L-202 | America | tropical japonica |
| aus-07 | BJ1 | India | aus | tej-01 | Koshihikari | Japan | temperate japonica |
| aus-08 | Kalamkati | India | aus | tej-02 | Geumobyeo | Korea | temperate japonica |
| aus-09 | Dhala Shaitta | Bangladesh | aus | tej-03 | Baghlani Nangarhar | Afghanistan | temperate japonica |
| aus-10 | Lalamon 158 | Bangladesh | aus | tej-04 | Aedal | Korea | temperate japonica |
| aus-11 | Aswina | Bangladesh | aus | tej-05 | TainanIku 487 | Taiwan | temperate japonica |
| aus-12 | Bamoia | Bangladesh | aus | tej-06 | KotobukiMochi | Japan | temperate japonica |
| ray-01 | Matia Amon | Bangladesh | rayada | tej-07 | Taipei 309 | Taiwan | temperate japonica |
| ray-02 | Kalamon 77-20 | Bangladesh | rayada | tej-08 | Chodongji | South Korea | temperate japonica |
| ray-03 | Jatra Motuk | Bangladesh | rayada | tej-09 | Beonjo | Korea | temperate japonica |
| ray-04 | Matiamon 53-13 | Bangladesh | rayada | tej-10 | Mansaku | Japan | temperate japonica |
| ray-05 | Khesail | Bangladesh | rayada | tej-11 | Oro | Chile | temperate japonica |
| ray-06 | DalKatra | Bangladesh | rayada | tej-12 | Luk Takhar | Afghanistan | temperate japonica |
| ray-07 | Lalamon 594 | Bangladesh | rayada | tej-13 | NepHoa Vang | Vietnam | temperate japonica |
| ray-08 | Nara Bet | Bangladesh | rayada |
表1 69份试验材料编号、名称、来源地及类型
Table 1 Serial number、name、origin and classification of rice accessions used in this study.
| 编号 Serial number | 品种名称 Accession name | 来源地 Country of origin | 类型 Classification | 编号 Serial number | 品种名称 Accession name | 来源地 Country of origin | 类型 Classification |
|---|---|---|---|---|---|---|---|
| ind-01 | Jaya | India | indica | aro-01 | JC101 | India | aromatic |
| ind-02 | IR8 | Phillipines | indica | aro-02 | Nga Ya Pauk | Myanmar | aromatic |
| ind-03 | Taichung Native 1 | Taiwan | indica | aro-03 | Kui Sali | Bangladesh | aromatic |
| ind-04 | Khao Dawk Mali 105 | Thailand | indica | aro-04 | DomSofid | Iran | aromatic |
| ind-05 | Padi Boenor | Indonesia | indica | aro-05 | Kalijira | Bangladesh | aromatic |
| ind-06 | JC92 | India | indica | aro-06 | Gordoi | Bangladesh | aromatic |
| ind-07 | Champa Tong 54 | Thailand | indica | aro-07 | JC73-4 | India | aromatic |
| ind-08 | Lageado | Brazil | indica | aro-08 | Shwewa | Myanmar | aromatic |
| ind-09 | Dhola Aman | Bangladesh | indica | aro-09 | Kanjari | India | aromatic |
| ind-10 | RTS12 | Vietnam | indica | aro-10 | Pankhari 203 | India | aromatic |
| ind-11 | IR36 | Phillipines | indica | aro-11 | N12 | India | aromatic |
| ind-12 | CO18 | India | indica | aro-12 | Basmati 1 | Pakistan | aromatic |
| ind-13 | Gie 57 | Vietnam | indica | trj-01 | Dou-Agu | Brazil | tropical japonica |
| ind-14 | Macan Binundok | Philippines | indica | trj-02 | Trembese | Indonesia | tropical japonica |
| ind-15 | TD25 | Thailand | indica | trj-03 | Gundil Kuning | Indonesia | tropical japonica |
| aus-01 | Shor Shori | Bangladesh | aus | trj-04 | Honduras | Brazil | tropical japonica |
| aus-02 | Jhona 349 | India | aus | trj-05 | Kinastano | Philippines | tropical japonica |
| aus-03 | Black Gora (Ncs12) | India | aus | trj-06 | Dholi Boro | Bangladesh | tropical japonica |
| aus-04 | Shada Boro | Bangladesh | aus | trj-07 | Cuba 65 | Cuba | tropical japonica |
| aus-05 | Unnamed | Bangladesh | aus | trj-08 | IAC25 | Brazil | tropical japonica |
| aus-06 | Badal 89 | Bangladesh | aus | trj-09 | L-202 | America | tropical japonica |
| aus-07 | BJ1 | India | aus | tej-01 | Koshihikari | Japan | temperate japonica |
| aus-08 | Kalamkati | India | aus | tej-02 | Geumobyeo | Korea | temperate japonica |
| aus-09 | Dhala Shaitta | Bangladesh | aus | tej-03 | Baghlani Nangarhar | Afghanistan | temperate japonica |
| aus-10 | Lalamon 158 | Bangladesh | aus | tej-04 | Aedal | Korea | temperate japonica |
| aus-11 | Aswina | Bangladesh | aus | tej-05 | TainanIku 487 | Taiwan | temperate japonica |
| aus-12 | Bamoia | Bangladesh | aus | tej-06 | KotobukiMochi | Japan | temperate japonica |
| ray-01 | Matia Amon | Bangladesh | rayada | tej-07 | Taipei 309 | Taiwan | temperate japonica |
| ray-02 | Kalamon 77-20 | Bangladesh | rayada | tej-08 | Chodongji | South Korea | temperate japonica |
| ray-03 | Jatra Motuk | Bangladesh | rayada | tej-09 | Beonjo | Korea | temperate japonica |
| ray-04 | Matiamon 53-13 | Bangladesh | rayada | tej-10 | Mansaku | Japan | temperate japonica |
| ray-05 | Khesail | Bangladesh | rayada | tej-11 | Oro | Chile | temperate japonica |
| ray-06 | DalKatra | Bangladesh | rayada | tej-12 | Luk Takhar | Afghanistan | temperate japonica |
| ray-07 | Lalamon 594 | Bangladesh | rayada | tej-13 | NepHoa Vang | Vietnam | temperate japonica |
| ray-08 | Nara Bet | Bangladesh | rayada |
| 引物 Marker | 染色体 Chromosome | 位置/(cM) Location /(cM) | 样本量 Sample | 等位 基因数 Allele | Nei基因 多样性指数 Nei's genetic diversity index | 引物 Marker | 染色体 Chromosome | 位置/(cM) Location /(cM) | 样本量 Sample | 等位 基因数 Allele | Nei基因 多样性指数 Nei's genetic diversity index |
|---|---|---|---|---|---|---|---|---|---|---|---|
| RM462 | 1 | 2.8 | 69 | 8 | 0.6704 | RM30 | 6 | 105.1 | 69 | 11 | 0.7146 |
| RM1195 | 1 | 30.5 | 69 | 11 | 0.7982 | RM340 | 6 | 113.1 | 69 | 17 | 0.8973 |
| RM583 | 1 | 43.2 | 69 | 12 | 0.8670 | RM176 | 6 | 120.9 | 69 | 4 | 0.6725 |
| RM490 | 1 | 51.0 | 69 | 5 | 0.7015 | RM481 | 7 | 17.6 | 69 | 20 | 0.9309 |
| RM449 | 1 | 73.1 | 69 | 9 | 0.8460 | RM180 | 7 | 42.2 | 69 | 11 | 0.7587 |
| RM129 | 1 | 75.9 | 69 | 5 | 0.7263 | RM542 | 7 | 49.7 | 69 | 13 | 0.8145 |
| RM488 | 1 | 102.3 | 69 | 13 | 0.8922 | RM418 | 7 | 61.8 | 69 | 11 | 0.8952 |
| RM443 | 1 | 121.0 | 69 | 4 | 0.7028 | RM432 | 7 | 64.7 | 69 | 4 | 0.6608 |
| RM297 | 1 | 132.0 | 69 | 13 | 0.8544 | RM336 | 7 | 77.8 | 69 | 19 | 0.9238 |
| RM212 | 1 | 135.8 | 69 | 7 | 0.7104 | RM505 | 7 | 84.4 | 69 | 7 | 0.5285 |
| RM472 | 1 | 146.4 | 69 | 14 | 0.8733 | RM234 | 7 | 93.9 | 69 | 12 | 0.8326 |
| RM414 | 1 | 161.8 | 69 | 2 | 0.4621 | RM18 | 7 | 94.7 | 69 | 7 | 0.8070 |
| RM485 | 2 | 0.0 | 69 | 15 | 0.8759 | RM420 | 7 | 118.3 | 69 | 5 | 0.7444 |
| RM555 | 2 | 20.0 | 69 | 4 | 0.6940 | RM337 | 8 | 0.5 | 69 | 9 | 0.7129 |
| RM8 | 2 | 26.9 | 69 | 6 | 0.5869 | RM25 | 8 | 35.2 | 69 | 6 | 0.7104 |
| RM423 | 2 | 28.7 | 69 | 8 | 0.7746 | RM72 | 8 | 45.4 | 69 | 14 | 0.8780 |
| RM71 | 2 | 40.9 | 69 | 16 | 0.8645 | RM331 | 8 | 54.3 | 69 | 14 | 0.7708 |
| RM475 | 2 | 92.5 | 69 | 11 | 0.8540 | RM404 | 8 | 55.4 | 69 | 15 | 0.8683 |
| RM263 | 2 | 104.9 | 69 | 9 | 0.7834 | RM515 | 8 | 72.2 | 69 | 15 | 0.8082 |
| RM208 | 2 | 154.1 | 69 | 15 | 0.9023 | RM210 | 8 | 86.7 | 69 | 10 | 0.8326 |
| RM498 | 2 | 157.1 | 69 | 4 | 0.6452 | RM149 | 8 | 101.1 | 69 | 10 | 0.8555 |
| RM132 | 3 | 2.5 | 69 | 4 | 0.5264 | RM230 | 8 | 105.7 | 69 | 6 | 0.7494 |
| RM231 | 3 | 11.5 | 69 | 8 | 0.7347 | RM477 | 8 | 120.4 | 69 | 2 | 0.4537 |
| RM545 | 3 | 25.0 | 69 | 13 | 0.8750 | RM316 | 9 | 0.8 | 69 | 10 | 0.8339 |
| RM232 | 3 | 44.3 | 69 | 15 | 0.8948 | RM219 | 9 | 20.7 | 69 | 15 | 0.8750 |
| RM7 | 3 | 44.4 | 69 | 6 | 0.7591 | RM321 | 9 | 40.1 | 69 | 2 | 0.4915 |
| RM554 | 3 | 55.3 | 69 | 5 | 0.7284 | RM409 | 9 | 49.3 | 69 | 4 | 0.4965 |
| RM411 | 3 | 87.1 | 69 | 5 | 0.6137 | RM566 | 9 | 50.7 | 69 | 17 | 0.8872 |
| RM426 | 3 | 122.3 | 69 | 23 | 0.9364 | RM257 | 9 | 64.1 | 69 | 23 | 0.9065 |
| RM448 | 3 | 140.1 | 69 | 14 | 0.8732 | RM278 | 9 | 74.7 | 69 | 14 | 0.8847 |
| RM293 | 3 | 142.3 | 69 | 4 | 0.6734 | RM215 | 9 | 83.2 | 69 | 13 | 0.8414 |
| RM422 | 3 | 151.5 | 69 | 13 | 0.8822 | RM205 | 9 | 93.5 | 69 | 24 | 0.9296 |
| RM85 | 3 | 166.4 | 69 | 11 | 0.7372 | RM474 | 10 | 3.0 | 69 | 19 | 0.9259 |
| RM551 | 4 | 3.1 | 69 | 16 | 0.8952 | RM216 | 10 | 12.5 | 69 | 10 | 0.8385 |
| RM5414 | 4 | 7.9 | 69 | 12 | 0.8532 | RM311 | 10 | 17.2 | 69 | 7 | 0.7664 |
| RM261 | 4 | 17.5 | 69 | 3 | 0.4243 | RM467 | 10 | 28.2 | 69 | 12 | 0.8691 |
| RM307 | 4 | 20.9 | 69 | 13 | 0.8456 | RM184 | 10 | 41.6 | 69 | 6 | 0.8087 |
| RM185 | 4 | 42.8 | 69 | 3 | 0.5268 | RM258 | 10 | 48.8 | 69 | 8 | 0.7356 |
| RM119 | 4 | 62.6 | 69 | 4 | 0.5480 | RM147 | 10 | 70.0 | 69 | 3 | 0.6011 |
| RM273 | 4 | 72.8 | 69 | 5 | 0.6268 | RM228 | 10 | 78.4 | 69 | 11 | 0.7940 |
| RM303 | 4 | 90.8 | 69 | 22 | 0.8960 | RM333 | 10 | 78.4 | 69 | 20 | 0.9355 |
| RM348 | 4 | 112.3 | 69 | 3 | 0.5138 | RM286 | 11 | 1.4 | 69 | 14 | 0.8876 |
| RM567 | 4 | 126.2 | 69 | 7 | 0.8309 | RM332 | 11 | 13.8 | 69 | 6 | 0.6356 |
| RM507 | 5 | 1.5 | 69 | 3 | 0.5150 | RM167 | 11 | 24.1 | 69 | 13 | 0.7893 |
| RM592 | 5 | 23.6 | 69 | 27 | 0.9452 | RM536 | 11 | 49.1 | 69 | 10 | 0.8465 |
| RM267 | 5 | 24.7 | 69 | 12 | 0.8158 | RM6091 | 11 | 56.2 | 69 | 7 | 0.7801 |
| RM169 | 5 | 48.6 | 69 | 10 | 0.6994 | RM209 | 11 | 69.4 | 69 | 15 | 0.8452 |
| RM5140 | 5 | 54.6 | 69 | 8 | 0.6499 | RM457 | 11 | 78.8 | 69 | 3 | 0.4192 |
| RM164 | 5 | 74.5 | 69 | 15 | 0.9044 | RM206 | 11 | 88.4 | 69 | 16 | 0.8809 |
| RM161 | 5 | 80.7 | 69 | 7 | 0.8208 | RM224 | 11 | 115.1 | 69 | 12 | 0.8687 |
| RM421 | 5 | 101.5 | 69 | 3 | 0.4318 | RM144 | 11 | 116.2 | 69 | 12 | 0.8456 |
| RM274 | 5 | 109.0 | 69 | 9 | 0.6906 | RM20A | 12 | 9.7 | 69 | 16 | 0.8263 |
| RM334 | 5 | 117.2 | 69 | 22 | 0.9162 | RM19 | 12 | 19.1 | 69 | 12 | 0.7742 |
| RM190 | 6 | 8.2 | 69 | 10 | 0.8423 | RM247 | 12 | 26.7 | 69 | 22 | 0.9032 |
| RM225 | 6 | 13.2 | 69 | 8 | 0.8158 | RM101 | 12 | 48.2 | 69 | 17 | 0.7478 |
| RM253 | 6 | 25.2 | 69 | 20 | 0.9095 | RM7102 | 12 | 51.8 | 69 | 8 | 0.7872 |
| RM527 | 6 | 56.3 | 69 | 14 | 0.8851 | RM511 | 12 | 60.3 | 69 | 5 | 0.5793 |
| RM5745 | 6 | 64.9 | 69 | 3 | 0.4814 | RM463 | 12 | 75.8 | 69 | 5 | 0.6767 |
| RM541 | 6 | 68.1 | 69 | 13 | 0.8368 | RM270 | 12 | 91.3 | 69 | 4 | 0.4058 |
| RM275 | 6 | 89.0 | 69 | 5 | 0.5238 | RM17 | 12 | 107.4 | 69 | 10 | 0.6822 |
表2 120对SSR引物的染色体位置及特征
Table 2 Chromosomal position and characters of the 120 SSR markers.
| 引物 Marker | 染色体 Chromosome | 位置/(cM) Location /(cM) | 样本量 Sample | 等位 基因数 Allele | Nei基因 多样性指数 Nei's genetic diversity index | 引物 Marker | 染色体 Chromosome | 位置/(cM) Location /(cM) | 样本量 Sample | 等位 基因数 Allele | Nei基因 多样性指数 Nei's genetic diversity index |
|---|---|---|---|---|---|---|---|---|---|---|---|
| RM462 | 1 | 2.8 | 69 | 8 | 0.6704 | RM30 | 6 | 105.1 | 69 | 11 | 0.7146 |
| RM1195 | 1 | 30.5 | 69 | 11 | 0.7982 | RM340 | 6 | 113.1 | 69 | 17 | 0.8973 |
| RM583 | 1 | 43.2 | 69 | 12 | 0.8670 | RM176 | 6 | 120.9 | 69 | 4 | 0.6725 |
| RM490 | 1 | 51.0 | 69 | 5 | 0.7015 | RM481 | 7 | 17.6 | 69 | 20 | 0.9309 |
| RM449 | 1 | 73.1 | 69 | 9 | 0.8460 | RM180 | 7 | 42.2 | 69 | 11 | 0.7587 |
| RM129 | 1 | 75.9 | 69 | 5 | 0.7263 | RM542 | 7 | 49.7 | 69 | 13 | 0.8145 |
| RM488 | 1 | 102.3 | 69 | 13 | 0.8922 | RM418 | 7 | 61.8 | 69 | 11 | 0.8952 |
| RM443 | 1 | 121.0 | 69 | 4 | 0.7028 | RM432 | 7 | 64.7 | 69 | 4 | 0.6608 |
| RM297 | 1 | 132.0 | 69 | 13 | 0.8544 | RM336 | 7 | 77.8 | 69 | 19 | 0.9238 |
| RM212 | 1 | 135.8 | 69 | 7 | 0.7104 | RM505 | 7 | 84.4 | 69 | 7 | 0.5285 |
| RM472 | 1 | 146.4 | 69 | 14 | 0.8733 | RM234 | 7 | 93.9 | 69 | 12 | 0.8326 |
| RM414 | 1 | 161.8 | 69 | 2 | 0.4621 | RM18 | 7 | 94.7 | 69 | 7 | 0.8070 |
| RM485 | 2 | 0.0 | 69 | 15 | 0.8759 | RM420 | 7 | 118.3 | 69 | 5 | 0.7444 |
| RM555 | 2 | 20.0 | 69 | 4 | 0.6940 | RM337 | 8 | 0.5 | 69 | 9 | 0.7129 |
| RM8 | 2 | 26.9 | 69 | 6 | 0.5869 | RM25 | 8 | 35.2 | 69 | 6 | 0.7104 |
| RM423 | 2 | 28.7 | 69 | 8 | 0.7746 | RM72 | 8 | 45.4 | 69 | 14 | 0.8780 |
| RM71 | 2 | 40.9 | 69 | 16 | 0.8645 | RM331 | 8 | 54.3 | 69 | 14 | 0.7708 |
| RM475 | 2 | 92.5 | 69 | 11 | 0.8540 | RM404 | 8 | 55.4 | 69 | 15 | 0.8683 |
| RM263 | 2 | 104.9 | 69 | 9 | 0.7834 | RM515 | 8 | 72.2 | 69 | 15 | 0.8082 |
| RM208 | 2 | 154.1 | 69 | 15 | 0.9023 | RM210 | 8 | 86.7 | 69 | 10 | 0.8326 |
| RM498 | 2 | 157.1 | 69 | 4 | 0.6452 | RM149 | 8 | 101.1 | 69 | 10 | 0.8555 |
| RM132 | 3 | 2.5 | 69 | 4 | 0.5264 | RM230 | 8 | 105.7 | 69 | 6 | 0.7494 |
| RM231 | 3 | 11.5 | 69 | 8 | 0.7347 | RM477 | 8 | 120.4 | 69 | 2 | 0.4537 |
| RM545 | 3 | 25.0 | 69 | 13 | 0.8750 | RM316 | 9 | 0.8 | 69 | 10 | 0.8339 |
| RM232 | 3 | 44.3 | 69 | 15 | 0.8948 | RM219 | 9 | 20.7 | 69 | 15 | 0.8750 |
| RM7 | 3 | 44.4 | 69 | 6 | 0.7591 | RM321 | 9 | 40.1 | 69 | 2 | 0.4915 |
| RM554 | 3 | 55.3 | 69 | 5 | 0.7284 | RM409 | 9 | 49.3 | 69 | 4 | 0.4965 |
| RM411 | 3 | 87.1 | 69 | 5 | 0.6137 | RM566 | 9 | 50.7 | 69 | 17 | 0.8872 |
| RM426 | 3 | 122.3 | 69 | 23 | 0.9364 | RM257 | 9 | 64.1 | 69 | 23 | 0.9065 |
| RM448 | 3 | 140.1 | 69 | 14 | 0.8732 | RM278 | 9 | 74.7 | 69 | 14 | 0.8847 |
| RM293 | 3 | 142.3 | 69 | 4 | 0.6734 | RM215 | 9 | 83.2 | 69 | 13 | 0.8414 |
| RM422 | 3 | 151.5 | 69 | 13 | 0.8822 | RM205 | 9 | 93.5 | 69 | 24 | 0.9296 |
| RM85 | 3 | 166.4 | 69 | 11 | 0.7372 | RM474 | 10 | 3.0 | 69 | 19 | 0.9259 |
| RM551 | 4 | 3.1 | 69 | 16 | 0.8952 | RM216 | 10 | 12.5 | 69 | 10 | 0.8385 |
| RM5414 | 4 | 7.9 | 69 | 12 | 0.8532 | RM311 | 10 | 17.2 | 69 | 7 | 0.7664 |
| RM261 | 4 | 17.5 | 69 | 3 | 0.4243 | RM467 | 10 | 28.2 | 69 | 12 | 0.8691 |
| RM307 | 4 | 20.9 | 69 | 13 | 0.8456 | RM184 | 10 | 41.6 | 69 | 6 | 0.8087 |
| RM185 | 4 | 42.8 | 69 | 3 | 0.5268 | RM258 | 10 | 48.8 | 69 | 8 | 0.7356 |
| RM119 | 4 | 62.6 | 69 | 4 | 0.5480 | RM147 | 10 | 70.0 | 69 | 3 | 0.6011 |
| RM273 | 4 | 72.8 | 69 | 5 | 0.6268 | RM228 | 10 | 78.4 | 69 | 11 | 0.7940 |
| RM303 | 4 | 90.8 | 69 | 22 | 0.8960 | RM333 | 10 | 78.4 | 69 | 20 | 0.9355 |
| RM348 | 4 | 112.3 | 69 | 3 | 0.5138 | RM286 | 11 | 1.4 | 69 | 14 | 0.8876 |
| RM567 | 4 | 126.2 | 69 | 7 | 0.8309 | RM332 | 11 | 13.8 | 69 | 6 | 0.6356 |
| RM507 | 5 | 1.5 | 69 | 3 | 0.5150 | RM167 | 11 | 24.1 | 69 | 13 | 0.7893 |
| RM592 | 5 | 23.6 | 69 | 27 | 0.9452 | RM536 | 11 | 49.1 | 69 | 10 | 0.8465 |
| RM267 | 5 | 24.7 | 69 | 12 | 0.8158 | RM6091 | 11 | 56.2 | 69 | 7 | 0.7801 |
| RM169 | 5 | 48.6 | 69 | 10 | 0.6994 | RM209 | 11 | 69.4 | 69 | 15 | 0.8452 |
| RM5140 | 5 | 54.6 | 69 | 8 | 0.6499 | RM457 | 11 | 78.8 | 69 | 3 | 0.4192 |
| RM164 | 5 | 74.5 | 69 | 15 | 0.9044 | RM206 | 11 | 88.4 | 69 | 16 | 0.8809 |
| RM161 | 5 | 80.7 | 69 | 7 | 0.8208 | RM224 | 11 | 115.1 | 69 | 12 | 0.8687 |
| RM421 | 5 | 101.5 | 69 | 3 | 0.4318 | RM144 | 11 | 116.2 | 69 | 12 | 0.8456 |
| RM274 | 5 | 109.0 | 69 | 9 | 0.6906 | RM20A | 12 | 9.7 | 69 | 16 | 0.8263 |
| RM334 | 5 | 117.2 | 69 | 22 | 0.9162 | RM19 | 12 | 19.1 | 69 | 12 | 0.7742 |
| RM190 | 6 | 8.2 | 69 | 10 | 0.8423 | RM247 | 12 | 26.7 | 69 | 22 | 0.9032 |
| RM225 | 6 | 13.2 | 69 | 8 | 0.8158 | RM101 | 12 | 48.2 | 69 | 17 | 0.7478 |
| RM253 | 6 | 25.2 | 69 | 20 | 0.9095 | RM7102 | 12 | 51.8 | 69 | 8 | 0.7872 |
| RM527 | 6 | 56.3 | 69 | 14 | 0.8851 | RM511 | 12 | 60.3 | 69 | 5 | 0.5793 |
| RM5745 | 6 | 64.9 | 69 | 3 | 0.4814 | RM463 | 12 | 75.8 | 69 | 5 | 0.6767 |
| RM541 | 6 | 68.1 | 69 | 13 | 0.8368 | RM270 | 12 | 91.3 | 69 | 4 | 0.4058 |
| RM275 | 6 | 89.0 | 69 | 5 | 0.5238 | RM17 | 12 | 107.4 | 69 | 10 | 0.6822 |
| 类群 Group | 等位基因数 Number of alleles | Nei基因多样性指数 Nei's genetic diversity index | ||||
|---|---|---|---|---|---|---|
| 最大 Maximum | 平均 Average | 最大 Maximum | 平均 Average | |||
| 总样本Total | 27 | 10.5 | 0.9452 | 0.7616 | ||
| indica | 9 | 4.3 | 0.8533 | 0.4990 | ||
| aus | 10 | 4.2 | 0.8889 | 0.5418 | ||
| rayada | 8 | 3.6 | 0.8750 | 0.4923 | ||
| aromatic | 10 | 4.4 | 0.8889 | 0.5311 | ||
| tropical japonica | 8 | 4.1 | 0.8642 | 0.5499 | ||
| temperate japonica | 11 | 4.2 | 0.8994 | 0.4851 | ||
表3 69份亚洲栽培稻分类遗传多样性特征
Table 3 Genetic diversity of the six sub-groups for 69 accessions of Oryza sativa L..
| 类群 Group | 等位基因数 Number of alleles | Nei基因多样性指数 Nei's genetic diversity index | ||||
|---|---|---|---|---|---|---|
| 最大 Maximum | 平均 Average | 最大 Maximum | 平均 Average | |||
| 总样本Total | 27 | 10.5 | 0.9452 | 0.7616 | ||
| indica | 9 | 4.3 | 0.8533 | 0.4990 | ||
| aus | 10 | 4.2 | 0.8889 | 0.5418 | ||
| rayada | 8 | 3.6 | 0.8750 | 0.4923 | ||
| aromatic | 10 | 4.4 | 0.8889 | 0.5311 | ||
| tropical japonica | 8 | 4.1 | 0.8642 | 0.5499 | ||
| temperate japonica | 11 | 4.2 | 0.8994 | 0.4851 | ||
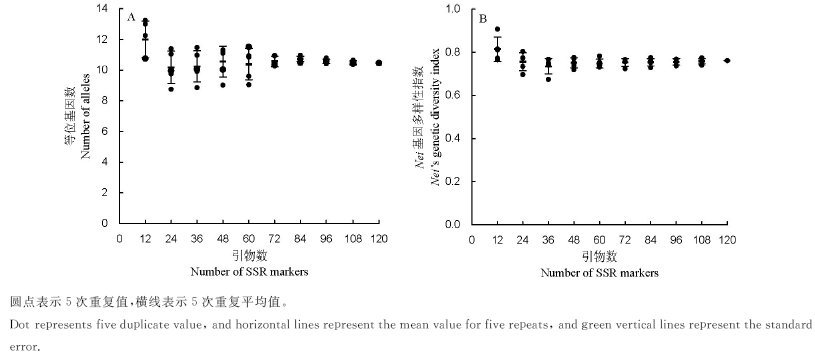
图1 69份试验材料不同SSR引物数下平均等位基因数(A)和Nei基因多样性指数(B)标准误
Fig. 1. Number of alleles (A) and Nei's genetic diversity index (B) with different numbers of SSR markers for 69 accessions.
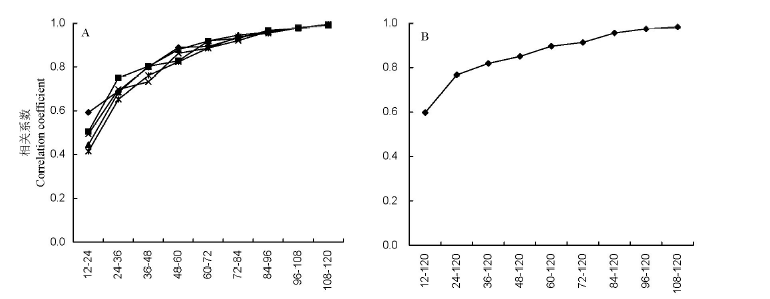
图2 69份试验材料相邻引物数遗传距离矩阵(A)及与母阵(B)的相关性测验
Fig. 2. Correlation test of genetic distance matrices between different numbers of markers (A) and 120 markers (B) for 69 accessions.
| [1] | 庄杰云, 施勇烽, 应杰政, 等. 中国主栽水稻品种微卫星标记数据库的初步构建. 中国水稻科学, 2006, 20(5): 460-468. |
| [2] | 李自超, 张洪亮, 曹永生. 中国地方稻种资源初级核心种质取样策略研究. 作物学报, 2003, 29: 20-24. |
| [3] | Gao L Z.Population structure and conservation genetics of wild rice Oryza rufipogon (Poaceae): A region-wide perspective from microsatellite variation.Mol Ecol, 2004, 13: 1009-1024. |
| [4] | Garris A J, Tai T H, Coburn J, et al.Genetic structure and diversity in Oryza sativa L.Genetics, 2005, 169: 1631-1638. |
| [5] | Li X B, Yan W G, Agrama H, et al.Genotypic and phenotypic characterization of genetic differentiation and diversity in the USDA rice mini-core collection.Genetics, 2010, 138: 1221-1230. |
| [6] | Pritchard J K, Rosenberg N A.Use of unlinked genetic markers to detect population stratification in association studies.Am J Hum Genet, 1999, 65: 220-228. |
| [7] | van Houdt J K J, Hellemans B, van de Putte A P, et al. Isolation and multiplex analysis of six polymorphic microsatellites in the Antarctic notothenioid fish,Trematomus newnesi. Mol Ecol Notes, 2006, 6: 157-159. |
| [8] | van de Putte A P, van Houdt J K J, et al. Species identification in the trematomid family using nuclear genetic markers.Polar Biol, 2009, 32: 1731-1741. |
| [9] | Zhang X Y, Li C W, Wang L F, et al.An estimation of the minimum number of SSR alleles needed to reveal genetic relationships in wheat varieties: Ⅰ. Information from large-scale planted varieties and cornerstone breeding parents in Chinese wheat improvement and production.Theor Appl Genet, 2002, 106: 112-117. |
| [10] | 王彪, 常汝镇, 陶莉, 等. 分析中国栽培大豆遗传多样性所需引物的数目. 分子植物育种, 2003, 1(1): 82-88. |
| [11] | 杨庆文, 陈成斌, 张万霞, 等. SSR等位变异数对普通野生稻居群遗传结构分析的影响. 中国水稻科学, 2005, 19(4): 297-302. |
| [12] | Glaszmann J C.Isozymes and classification of Asian rice varieties.Theor Appl Genet, 1987, 74: 21-30. |
| [13] | Wang C H, Zheng X M, Xu Q, et al.Genetic diversity and classification of Oryza sativa with emphasis on Chinese rice germplasm.Heredity, 2014, 112: 489-496. |
| [14] | Zheng K L, Subudi P K, Domingo J, et al.Rapid DNA isolation for marker assisted selection in rice breeding.Rice Genet News, 1995, 12: 255-258. |
| [15] | Temnykh S, Park W D, Ayres N, et al.Mapping and genome organization of microsatellite sequences in rice (Oryza sativa L.).Theor Appl Genet, 2000, 100: 697-712. |
| [16] | McCouch S R, Teytelman L, Xu Y B, et al. Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.).DNA Res, 2002, 9: 199-207. |
| [17] | 王彩红, 徐群, 于萍, 等. 亚洲栽培稻程氏指数与SSR标记分类的比较分析. 中国水稻科学, 2012, 26(2): 165-172. |
| [18] | Nei M, Li W H.Mathematical model for studying genetic variation in terms of restriction endonucleases.Proc Nat Acad Sci USA, 1979, 76: 5269-5273. |
| [19] | Mantel N A.The detection of disease clustering and a generalized regression approach.Cancer Res, 1967, 27: 209-220. |
| [20] | Matsuo T.Gen-ecological studies on cultivated rice.Bull Natl Inst Agric Sci, 1952, D3: 1-111. (in Japanese) |
| [21] | Banks M A,Eichert W, Olsen J B.Which genetic loci have greater population assignment power?Bioinformatics, 2003, 19: 143-1438. |
| [22] | Agrama H A, McClung A M, Yan W G. Using minimum DNA marker loci for accurate population classification in rice (Oryza sativa L.).Mol Breeding, 2012, 413-425. |
| [1] | 吴玥, 梁铖玮, 赵辰妃, 孙健, 马殿荣. 直播稻田杂草稻灾害发生及生态型的演变特征[J]. 中国水稻科学, 2024, 38(4): 447-455. |
| [2] | 董俊杰, 曾宇翔, 季芝娟, 梁燕, 杨长登. 273份水稻种质资源的遗传多样性、群体结构与连锁不平衡 分析[J]. 中国水稻科学, 2021, 35(2): 130-140. |
| [3] | 朱名海, 彭丹丹, 舒灿伟, 周而勋. 海南南繁区水稻纹枯病菌的遗传多样性与致病力分化[J]. 中国水稻科学, 2019, 33(2): 176-185. |
| [4] | 朱名海, 皮磊, 舒灿伟, 周而勋. 南繁区稻瘟病菌遗传多样性和群体遗传结构的AFLP分析[J]. 中国水稻科学, 2017, 31(3): 320-326. |
| [5] | 王玲, 左示敏, 张亚芳, 陈宗祥, 潘学彪, 黄世文. 四川省稻瘟病菌群体遗传结构分析[J]. 中国水稻科学, 2015, 29(3): 327-334. |
| [6] | 刘承晨, 赵富伟, 吴晓霞, 张昌泉, 朱孔志, 薛达元, 武建勇, 黄绍文, 徐小颖, 金银根, 刘巧泉. 云南哈尼梯田当前栽培水稻遗传多样性及群体结构分析[J]. 中国水稻科学, 2015, 29(1): 28-34. |
| [7] | 刘二宝1,刘洋1,刘晓丽1,刘强明1,赵凯铭1,2,EDZESI Wisdom Mawuli1 ,洪德林1,*. 杂交粳稻亲本产量性状优异配合力的标记基因型检测[J]. 中国水稻科学, 2013, 27(5): 473-481. |
| [8] | 牛付安1,2 ,陈兰1 ,张红1 ,袁勤2 ,程灿2 ,周继华2 ,洪德林1,*. 粳稻穗角性状优异等位变异的挖掘[J]. 中国水稻科学, 2013, 27(4): 373-380. |
| [9] | 孙建昌1,2,余滕琼3,汤翠凤3,曹桂兰1,徐福荣3,韩龙植1,*. 基于SSR标记的云南地方稻种群体内遗传多样性分析[J]. 中国水稻科学, 2013, 27(1): 41-48. |
| [10] | 周振玲1,2,王宝祥1,樊继伟1,卢百关1,赵志刚2,江玲2,秦德荣1,万建民2,3 ,徐大勇1,2,*. 江淮稻区不同生态型粳稻品种的籼粳分化度和遗传多样性[J]. 中国水稻科学, 2012, 26(4): 431-437. |
| [11] | 徐建欣,王云月*,姚春,刘云霞,汤淼,陆明兴. 利用SSR分子标记分析云南陆稻品种遗传多样性[J]. 中国水稻科学, 2012, 26(2): 155-164. |
| [12] | 王彩红,徐群,于萍,袁筱萍,余汉勇,王一平,汤圣祥,魏兴华*. 亚洲栽培稻程氏指数与SSR标记分类的比较分析[J]. 中国水稻科学, 2012, 26(2): 165-172. |
| [13] | 刘丹,孙健,马殿荣*,王嘉宇,唐亮,苗微,陈温福*. 利用SRAP分子标记分析47份杂草稻样品遗传多样性[J]. 中国水稻科学, 2012, 26(1): 70-76. |
| [14] | 张荣胜1,2,陈志谊1,*,刘永锋1. 水稻细菌性条斑病菌遗传多样性和致病型分化研究[J]. 中国水稻科学, 2011, 25(5): 523-528. |
| [15] | 董超, 徐福荣, 杨文毅, 张恩来, 杨雅云, 汤翠凤, 阿新祥, 戴陆园,. 布朗族当前种植稻作地方品种的SSR位点多样性分析[J]. 中国水稻科学, 2011, 25(2): 175-181 . |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||