
Chinese Journal OF Rice Science ›› 2025, Vol. 39 ›› Issue (6): 813-824.DOI: 10.16819/j.1001-7216.2025.241003
• Research Papers • Previous Articles Next Articles
LU Shuai1, TAO Tao1, LIU Ran1, ZHOU Wenyu1, CAO Lei1, YANG Qingqing1, ZHANG Mingqiu1, REN Xinzhe1, YANG Zhidi1, XU Fuxiang1, HUAN Haidong1, GONG Yuanhang1, ZHANG Haocheng1, JIN Sukui3, CAI Xiuling1,2, GAO Jiping1,2, LENG Yujia1,2,*( )
)
Received:2024-10-11
Revised:2024-12-02
Online:2025-11-10
Published:2025-11-19
Contact:
LENG Yujia
陆帅1, 陶涛1, 刘冉1, 周文玉1, 曹蕾1, 杨青青1, 张明秋1, 任鑫哲1, 杨芝笛1, 徐福祥1, 环海东1, 龚远航1, 张皓程1, 金素奎3, 蔡秀玲1,2, 高继平1,2, 冷语佳1,2,*( )
)
通讯作者:
冷语佳
基金资助:LU Shuai, TAO Tao, LIU Ran, ZHOU Wenyu, CAO Lei, YANG Qingqing, ZHANG Mingqiu, REN Xinzhe, YANG Zhidi, XU Fuxiang, HUAN Haidong, GONG Yuanhang, ZHANG Haocheng, JIN Sukui, CAI Xiuling, GAO Jiping, LENG Yujia. Identification and Gene Cloning of a Long Sterile Lemma and Small Grain Mutant lsg8 in Rice (Oryza sativa L.)[J]. Chinese Journal OF Rice Science, 2025, 39(6): 813-824.
陆帅, 陶涛, 刘冉, 周文玉, 曹蕾, 杨青青, 张明秋, 任鑫哲, 杨芝笛, 徐福祥, 环海东, 龚远航, 张皓程, 金素奎, 蔡秀玲, 高继平, 冷语佳. 水稻长护颖小粒突变体lsg8的表型鉴定与基因克隆[J]. 中国水稻科学, 2025, 39(6): 813-824.
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.ricesci.cn/EN/10.16819/j.1001-7216.2025.241003
| 引物名称 Primer name | 物理位置 Physical location(bp) | 正向引物序列 Forward primer sequence | 反向引物序列 Reverse primer sequence |
|---|---|---|---|
| M1 | 201350 | GTTTGAACAGTAGGACTTGT | AGAACATCTCACACTTCTCT |
| M2 | 4588509 | ATCTCCCTCCCTCTCCTCAC | TCCACACCTTCACAGTTGAC |
| M3 | 1135865 | TGGGATAGGAGTAGCACTTTTGG | CCATACATTCCAAACCATCCTAG |
| M4 | 2372755 | GCAAACTGTGAGCAACAATGG | AAACAAGACCATGCTCGTCGG |
| M5 | 3502364 | AATTTTACACCGGATCTAAACAC | ATGGAAATGCAAATTAAGAACAC |
| M6 | 3778634 | GTTCTGTTTCTTGCCCGACCTTT | GAATATGACCCACATGCCGACTC |
| M7 | 4092309 | ATCTAAAGGAGATCGGATGGTAT | AAGCATCCAGAGGTCGCAGCAAA |
Table 1. Primers and sequences used in gene mapping
| 引物名称 Primer name | 物理位置 Physical location(bp) | 正向引物序列 Forward primer sequence | 反向引物序列 Reverse primer sequence |
|---|---|---|---|
| M1 | 201350 | GTTTGAACAGTAGGACTTGT | AGAACATCTCACACTTCTCT |
| M2 | 4588509 | ATCTCCCTCCCTCTCCTCAC | TCCACACCTTCACAGTTGAC |
| M3 | 1135865 | TGGGATAGGAGTAGCACTTTTGG | CCATACATTCCAAACCATCCTAG |
| M4 | 2372755 | GCAAACTGTGAGCAACAATGG | AAACAAGACCATGCTCGTCGG |
| M5 | 3502364 | AATTTTACACCGGATCTAAACAC | ATGGAAATGCAAATTAAGAACAC |
| M6 | 3778634 | GTTCTGTTTCTTGCCCGACCTTT | GAATATGACCCACATGCCGACTC |
| M7 | 4092309 | ATCTAAAGGAGATCGGATGGTAT | AAGCATCCAGAGGTCGCAGCAAA |
| 引物名称 Primer name | 正向引物序列 Forward primer sequence | 反向引物序列 Reverse primer sequence |
|---|---|---|
| CL-1 | CCTCATCCTGCAGTTCCTCG | AACCTCAGCATATTTGGCGT |
| CL-2 | ATGGGCATGCTCCAACCAAT | GAGCCAACTTCCCATATCCCA |
| CL-3 | TCTTTGTCCATGTTCTGTCCACT | CGCATTGCAAATTAAAACGGCA |
| CL-4 | TGAAGGTCACGAAGCTCCAG | ATTTGTTGAGGTGGCCCTCG |
| CL-5 | TAATTTTGCAGGCGAGCCCA | CGTTGCCATTTCCACAGCTT |
| CL-6 | AGGCTGTTCAGTTCTGAGCAATA | TATAGTCCACTTGCCACCGC |
| CL-7 | TGCTCAGTGACCTTCAACTAACT | CCCCACTTTGGGTCTGAGTC |
Table 2. Primer sequences used for LSG8 gene sequencing
| 引物名称 Primer name | 正向引物序列 Forward primer sequence | 反向引物序列 Reverse primer sequence |
|---|---|---|
| CL-1 | CCTCATCCTGCAGTTCCTCG | AACCTCAGCATATTTGGCGT |
| CL-2 | ATGGGCATGCTCCAACCAAT | GAGCCAACTTCCCATATCCCA |
| CL-3 | TCTTTGTCCATGTTCTGTCCACT | CGCATTGCAAATTAAAACGGCA |
| CL-4 | TGAAGGTCACGAAGCTCCAG | ATTTGTTGAGGTGGCCCTCG |
| CL-5 | TAATTTTGCAGGCGAGCCCA | CGTTGCCATTTCCACAGCTT |
| CL-6 | AGGCTGTTCAGTTCTGAGCAATA | TATAGTCCACTTGCCACCGC |
| CL-7 | TGCTCAGTGACCTTCAACTAACT | CCCCACTTTGGGTCTGAGTC |
| 基因 Gene | 正向引物序列 Forward primer sequence | 反向引物序列 Reverse primer sequence |
|---|---|---|
| OsMADS34 | AGGAATATGTGAACTTGAA | TTCTGACTACTTGACTCT |
| SAD1 | CCAGGGGAGAAATCCAAGA | CTGTCGACCAAGCTTCAGG |
| LRG1 | ATGGAGTTTGGGATGGTGGAG | CAGCTTGTGCGCGTTCTGGT |
| EG1 | AACGTACACGACCCGATCAC | GACGTGGGTGTAGCAGGAGT |
| ASP1 | TCTTGTTCAACCTCCAAACACAGC | CAATGGCAGGAGCACTTGTTGG |
| OsVIL2 | GGAGTATGCTTTCCGGATCA | GTGGGAAACAACATGTGCAG |
| OsIG1 | TTCATCAACGTCGGGCACT | CTCCCCTTCGTAGCTCCTC |
| SNB | ACCACGAAGTAGGGAACGACTGGG | CAGCCAATAAGTCCTCAGTGGCCTG |
| OsIDS1 | CTGGCCTCCAGTTAACTTGT | GGCGCCGGCAGAGAATCCT |
| MFS1 | CGGCTCGTGATCTCGACACGTAC | CACAGCCGGACCAGTGCTCTC |
| LSG8 | TCTTGTTCAACCTCCAAACACAGC | CAATGGCAGGAGCACTTGTTGG |
| OsMADS1 | CCAAGCCACTCTTCTTGTTCG | TGATGGTGAGCATGAGGGTG |
| OsMADS6 | CCAACAATGCACTTTCTGAAAC | GGAGGCTTGCTGCATGGC |
| OsMADS14 | CCATTAACGAGCTTCAACGG | TGGTATGGATCTGAAGCCTCC |
| OsMADS15 | AGTACGCCACTGACTCCAGG | TGCTGGCCCCTCACATTC |
| DL | CCCATCTGCTTACAACCGCTT | GTTGGAGGTGGAAACCGTCG |
| OsEXPA1 | TGCAGAGCCTCCAATAGTAGTCCA | GGTACATCAAGCCTCTGTAGTGCAA |
| OsEXPB5 | TGTTTGTTAACGTCGCCGCGATAG | TCACTAGAAGCAGCTCTGCAAACG |
| OsEXPA10 | TCTTGTGCTCGTGACAAACGTTGC | CATTGGCATCCAGTCGGTTGAGTT |
| OsEXPB11 | GCAGTGCAGAGTTGCGGTAAATTG | ATCGACGACGACACAGTCACATCA |
| OsEXPA25 | TGGATCACGCTGAACCGGAACT | TGTAGATGTAGAGCGTCTGGCCG |
| UBQ | ACCACTTCGACCGCCACTACT | ACGCCTAAGCCTGCTGGTT |
Table 3. Primers for RT-qPCR
| 基因 Gene | 正向引物序列 Forward primer sequence | 反向引物序列 Reverse primer sequence |
|---|---|---|
| OsMADS34 | AGGAATATGTGAACTTGAA | TTCTGACTACTTGACTCT |
| SAD1 | CCAGGGGAGAAATCCAAGA | CTGTCGACCAAGCTTCAGG |
| LRG1 | ATGGAGTTTGGGATGGTGGAG | CAGCTTGTGCGCGTTCTGGT |
| EG1 | AACGTACACGACCCGATCAC | GACGTGGGTGTAGCAGGAGT |
| ASP1 | TCTTGTTCAACCTCCAAACACAGC | CAATGGCAGGAGCACTTGTTGG |
| OsVIL2 | GGAGTATGCTTTCCGGATCA | GTGGGAAACAACATGTGCAG |
| OsIG1 | TTCATCAACGTCGGGCACT | CTCCCCTTCGTAGCTCCTC |
| SNB | ACCACGAAGTAGGGAACGACTGGG | CAGCCAATAAGTCCTCAGTGGCCTG |
| OsIDS1 | CTGGCCTCCAGTTAACTTGT | GGCGCCGGCAGAGAATCCT |
| MFS1 | CGGCTCGTGATCTCGACACGTAC | CACAGCCGGACCAGTGCTCTC |
| LSG8 | TCTTGTTCAACCTCCAAACACAGC | CAATGGCAGGAGCACTTGTTGG |
| OsMADS1 | CCAAGCCACTCTTCTTGTTCG | TGATGGTGAGCATGAGGGTG |
| OsMADS6 | CCAACAATGCACTTTCTGAAAC | GGAGGCTTGCTGCATGGC |
| OsMADS14 | CCATTAACGAGCTTCAACGG | TGGTATGGATCTGAAGCCTCC |
| OsMADS15 | AGTACGCCACTGACTCCAGG | TGCTGGCCCCTCACATTC |
| DL | CCCATCTGCTTACAACCGCTT | GTTGGAGGTGGAAACCGTCG |
| OsEXPA1 | TGCAGAGCCTCCAATAGTAGTCCA | GGTACATCAAGCCTCTGTAGTGCAA |
| OsEXPB5 | TGTTTGTTAACGTCGCCGCGATAG | TCACTAGAAGCAGCTCTGCAAACG |
| OsEXPA10 | TCTTGTGCTCGTGACAAACGTTGC | CATTGGCATCCAGTCGGTTGAGTT |
| OsEXPB11 | GCAGTGCAGAGTTGCGGTAAATTG | ATCGACGACGACACAGTCACATCA |
| OsEXPA25 | TGGATCACGCTGAACCGGAACT | TGTAGATGTAGAGCGTCTGGCCG |
| UBQ | ACCACTTCGACCGCCACTACT | ACGCCTAAGCCTGCTGGTT |
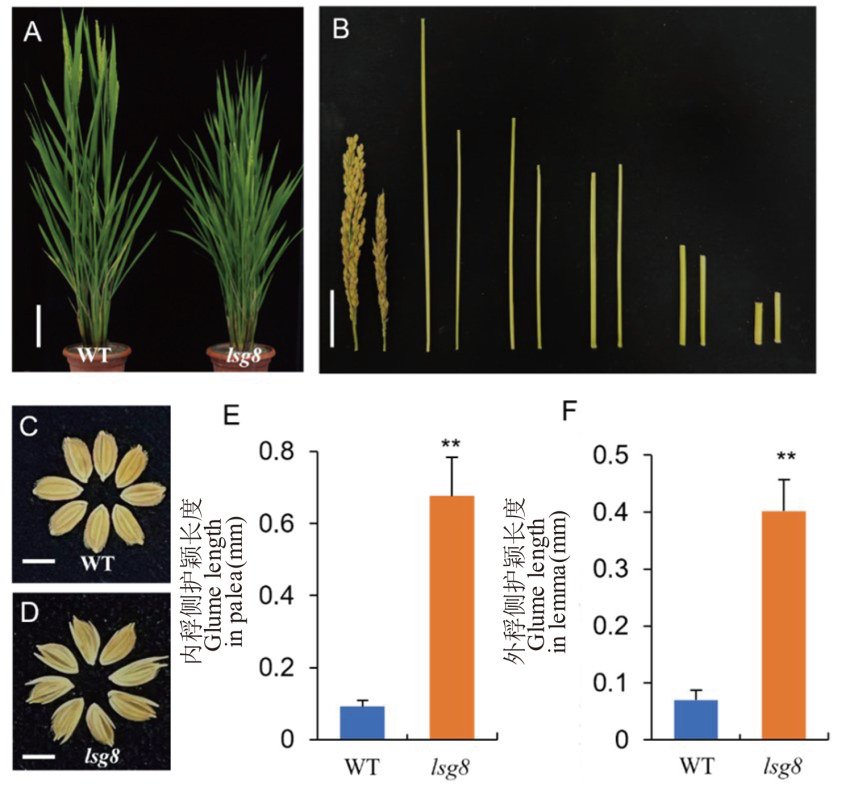
Fig. 1. Comparison of phenotypic traits between the wild type WYJ27 and lsg8 A, Phenotypic comparison of the wild type WYJ27 and lsg8 at heading stage, scale bar = 10 cm. B, Comparison of spikelet and internode length between the wild type WYJ27 and lsg8. The left is the wild type WYJ27, and the right is lsg8, scale bar=5 cm. C, Mature seeds of wild type WYJ27, scale bar=5 cm. D, Mature seeds of lsg8, scale bar=5 cm. E, Comparison of glume length on palea between wild type WYJ27 and lsg8. F, Comparison of glume length on lemma between wild type WYJ27 and lsg8. The t-test was used for significance analysis, and ** means significant difference at P < 0.01.
| 性状 Trait | WYJ27 | lsg8 |
|---|---|---|
| 株高 Plant height (cm) | 89.1 ± 2.8 | 76.8 ± 1.3** |
| 穗长 Panicle length (cm) | 17.0 ± 1.2 | 15.2 ± 0.9** |
| 倒1节长The 1st internode from the top (cm) | 26.9 ± 1.6 | 19.5 ± 1.4** |
| 倒2节长The 2nd internode from the top (cm) | 19.7 ± 0.8 | 17.6 ± 0.2** |
| 倒3节长The 3rd internode from the top (cm) | 12.9 ± 0.9 | 13.0 ± 1.2 |
| 倒4节长The 4th internode from the top (cm) | 9.4 ± 1.0 | 8.4 ± 1.0* |
| 倒5节长The 5th internode from the top (cm) | 4.3 ± 1.6 | 3.5 ± 1.1 |
| 分蘖数 No. of tillers per plant | 7.4 ± 1.4 | 16.4 ± 2.9** |
| 一次枝梗数 No. of primary rachis branches per panicle | 13.9 ± 1.0 | 10.1 ± 2.3** |
| 二次枝梗数No. of secondary rachis branches per panicle | 35.8 ± 5.4 | 24.9 ± 6.8** |
| 每穗粒数No. of spikelets per panicle | 208.1 ± 14.1 | 115.9 ± 8.9** |
| 结实率 Seed-setting rate (%) | 93.8 ± 2.3 | 61.5 ± 9.9** |
Table 4. Comparison of agronomic traits between WYJ27 and lsg8
| 性状 Trait | WYJ27 | lsg8 |
|---|---|---|
| 株高 Plant height (cm) | 89.1 ± 2.8 | 76.8 ± 1.3** |
| 穗长 Panicle length (cm) | 17.0 ± 1.2 | 15.2 ± 0.9** |
| 倒1节长The 1st internode from the top (cm) | 26.9 ± 1.6 | 19.5 ± 1.4** |
| 倒2节长The 2nd internode from the top (cm) | 19.7 ± 0.8 | 17.6 ± 0.2** |
| 倒3节长The 3rd internode from the top (cm) | 12.9 ± 0.9 | 13.0 ± 1.2 |
| 倒4节长The 4th internode from the top (cm) | 9.4 ± 1.0 | 8.4 ± 1.0* |
| 倒5节长The 5th internode from the top (cm) | 4.3 ± 1.6 | 3.5 ± 1.1 |
| 分蘖数 No. of tillers per plant | 7.4 ± 1.4 | 16.4 ± 2.9** |
| 一次枝梗数 No. of primary rachis branches per panicle | 13.9 ± 1.0 | 10.1 ± 2.3** |
| 二次枝梗数No. of secondary rachis branches per panicle | 35.8 ± 5.4 | 24.9 ± 6.8** |
| 每穗粒数No. of spikelets per panicle | 208.1 ± 14.1 | 115.9 ± 8.9** |
| 结实率 Seed-setting rate (%) | 93.8 ± 2.3 | 61.5 ± 9.9** |
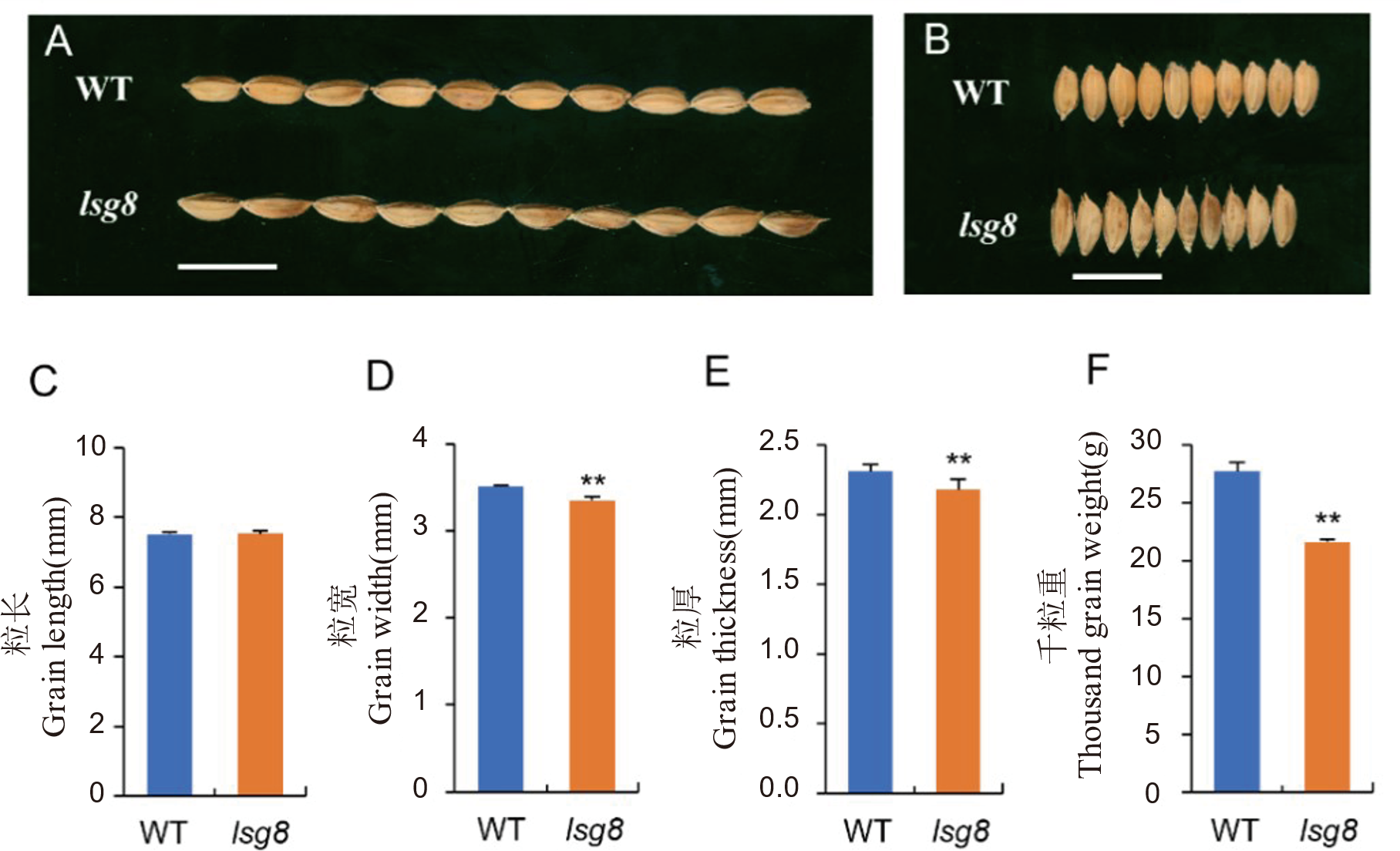
Fig. 2. Comparison of grain size between the wild type WYJ27 and lsg8 A, Comparison of grain length between the wild type WYJ27 and the lsg8 mutant, scale bar=1 cm. B, Comparison of grain width between the wild type WYJ27 and lsg8, scale bar=0.5 cm. C-F, Comparison of grain length (C), grain size (D), grain thickness (E) and 1000-grain weight (F) between the wild type WYJ27 and lsg8. The t-test was used for significance analysis. ** means significant difference at P < 0.01.
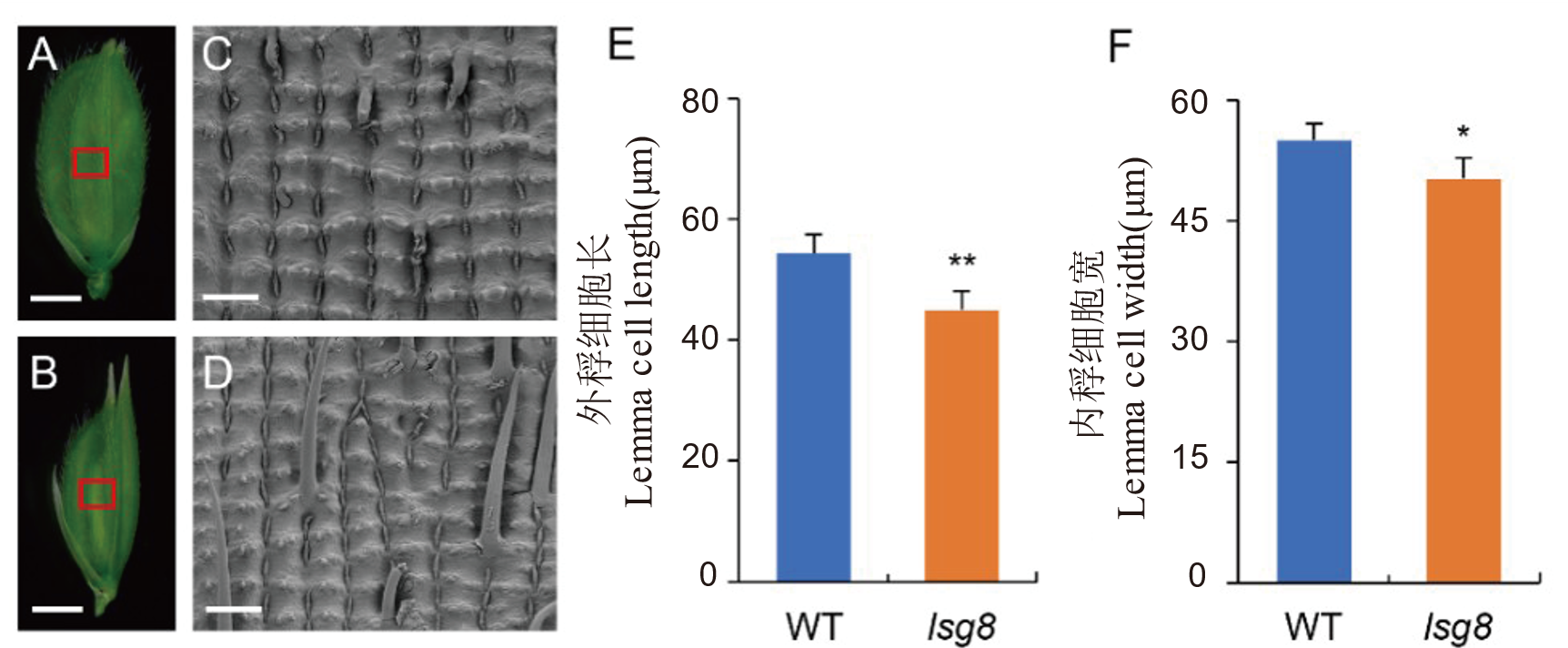
Fig. 3. Scanning electron microscopy of epidermal cells of the glume in wild type WYJ27 and the lsg8 mutant A, Glume of wild type WYJ27, scale bar=2 mm. B, Glume of mutant lsg8, scale bar= 2 mm. C, Outer epidermal cells of wild type WYJ27 glume, scale bar=100 μm. D, Outer epidermal cells of mutant lsg8 glume, scale bar=100 μm. E, Comparison of lemma cell length between wild type WYJ27 and lsg8. F, Comparison of lemma cell width between wild type WYJ27 and lsg8. Significance analysis was performed using t-test. * P<0.05; ** P<0.01.
| 杂交组合 Cross | F1 | F2 | χ2 (3:1) | |||
|---|---|---|---|---|---|---|
| 正常表型 Wild type | 突变表型 Mutant | 正常表型 Wild type | 突变表型 Mutant | |||
| lsg8 /IR36 | 15 | 0 | 1253 | 439 | 1.1046 | |
| IR36/ lsg8 | 23 | 0 | 856 | 295 | 0.2436 | |
Table 5. Genetic analysis of lsg8 mutant
| 杂交组合 Cross | F1 | F2 | χ2 (3:1) | |||
|---|---|---|---|---|---|---|
| 正常表型 Wild type | 突变表型 Mutant | 正常表型 Wild type | 突变表型 Mutant | |||
| lsg8 /IR36 | 15 | 0 | 1253 | 439 | 1.1046 | |
| IR36/ lsg8 | 23 | 0 | 856 | 295 | 0.2436 | |
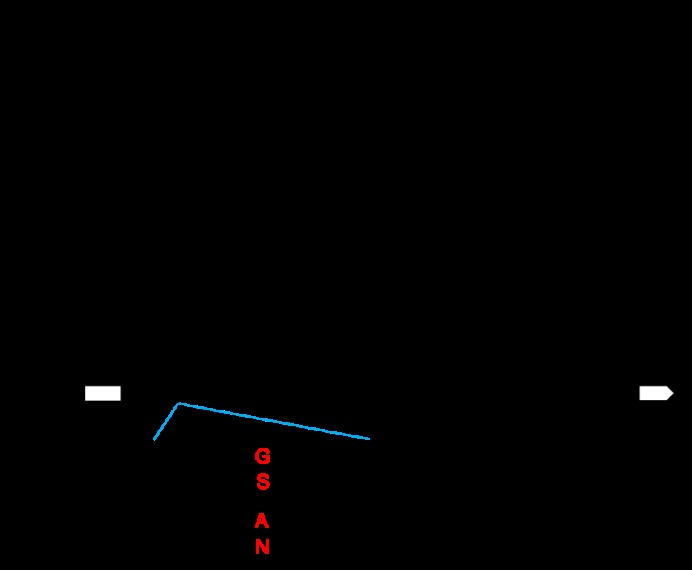
Fig. 4. Gene mapping of LSG8 A, Mapping of LSG8 on chromosome 8. B, Candidate gene LSG8. The red letters indicate the differences in bases and encoded amino acids between the wild type and the mutant on the candidate gene LSG8.
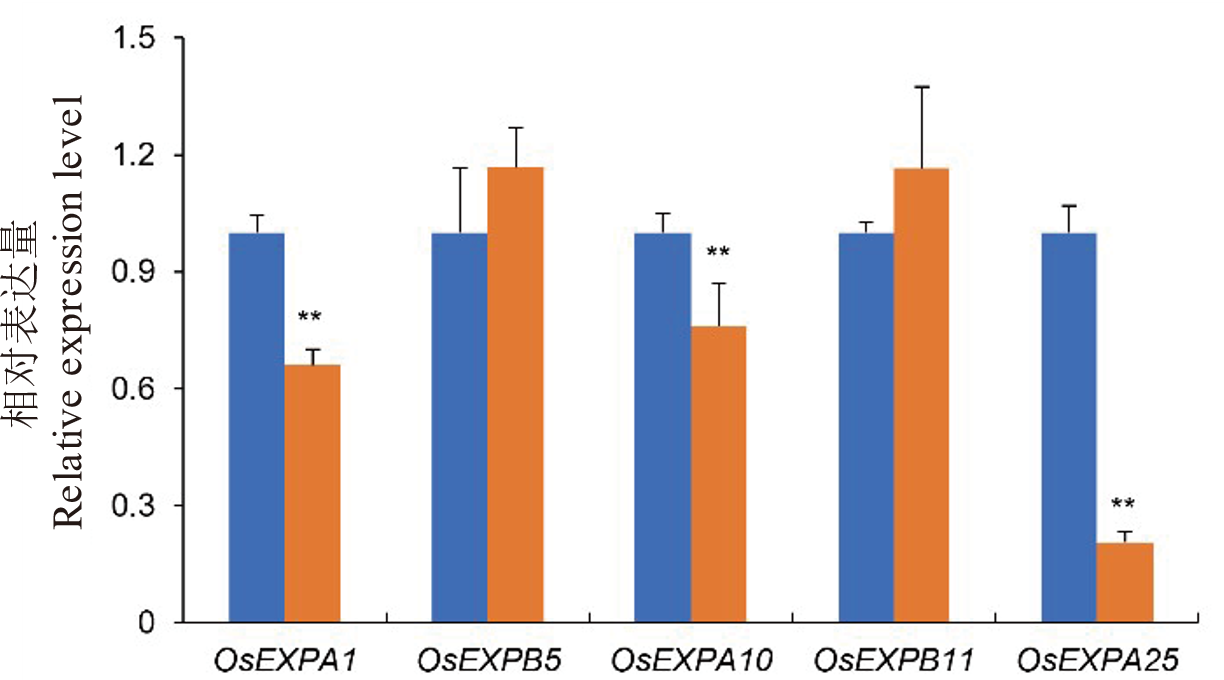
Fig. 6. Relative expression levels of cell expansins related genes in wild type WYJ27 and lsg8 mutant Each sample was analyzed in three independent biological replicates. ** Significant difference at P<0.01 by t-test, respectively.
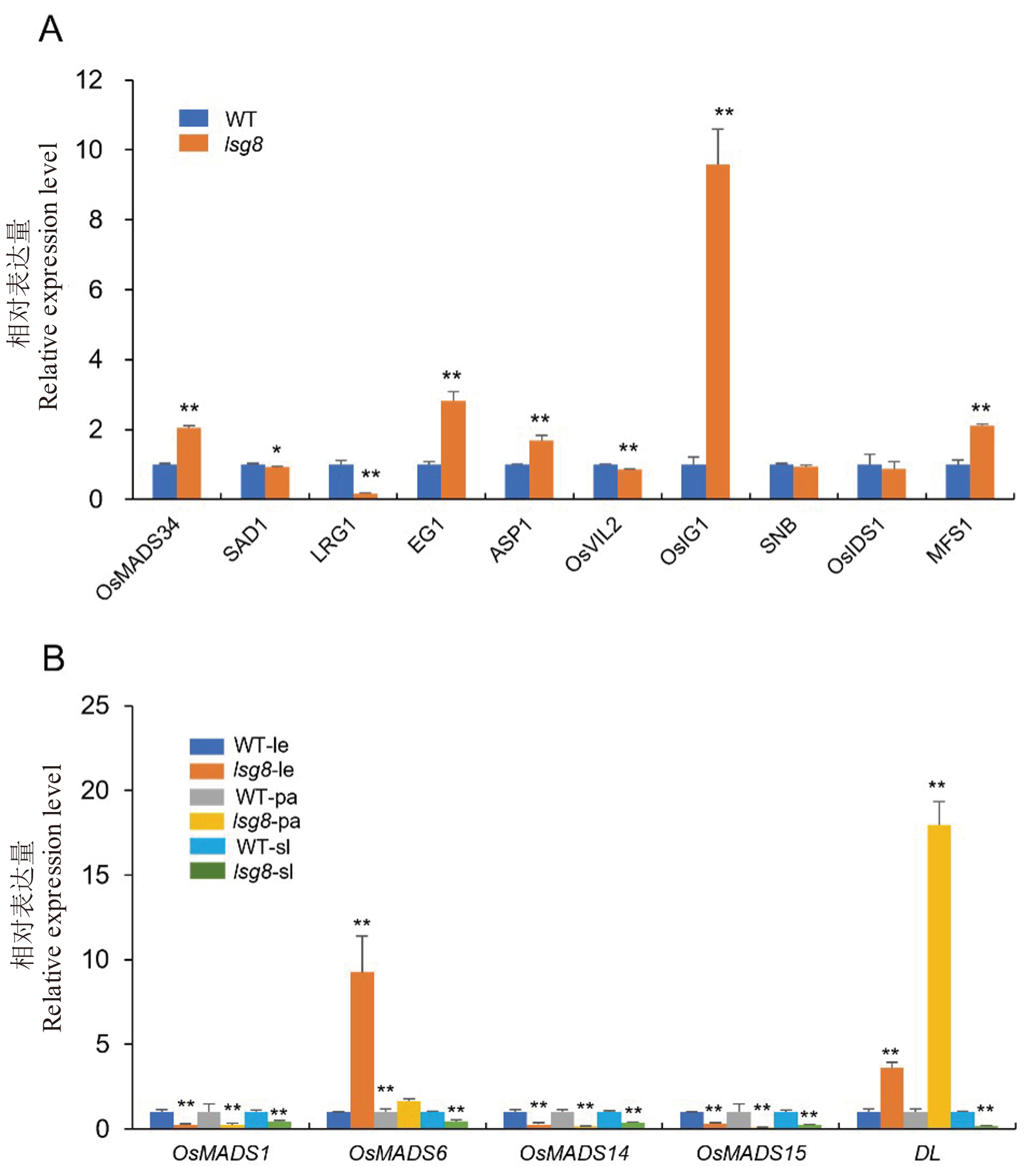
Fig. 7. Relative expression levels of genes regulating sterile lemma development and hull characteristics in wild type WYJ27 and the lsg8 mutant A, qPCR analysis of sterile lemma development regulatory genes (OsMADS34, SAD1, LRG1, EG1, ASP1, OsVIL2, OsIG1, SNB, OsIDS1 and MFS’) in the wild type and lsg8 mutant. B, qPCR analysis of floral organ identity genes (OsMADS1, OsMADS6, OsMADS14, OsMADS15, and DL) in the wild type and lsg8 mutant. le, Lemma; pa, Palea; sl, Sterile lemma. Each sample was analyzed in three independent biological replicates. * and ** : Significant difference at P<0.05 and P<0.01 by t-test, respectively.
| [1] | 朱玲, 陈晓琼, 杜康兮, 韩保林, 冉秀华, 张红宇, 徐培洲, 吴先军. 水稻长护颖突变体基因的克隆与表达分析[J]. 中国农业科学, 2015, 48(11): 2085-2095. |
| Zhu L, Chen X Q, Du K X, Han B L, Ran X H, Zhang H Y, Xu P Z, Wu X J. Gene cloning and expression analysis of long empty glumes mutants in rice[J]. Scientia Agricultura Sinica, 2015, 48(11): 2085-2095. (in Chinese with English abstract) | |
| [2] | 符德保, 李燕, 肖景华, 张启发, 吴昌银. 中国水稻基因组学研究历史及现状[J]. 生命科学, 2016, 28(10): 1113-1121. |
| Fu D B, Li Y, Xiao J H, Zhang Q F, Wu C Y. The history and current status of rice genomics research in China[J]. Chinese Bulletin of Life Sciences, 2016, 28(10): 1113-1121. (in Chinese with English abstract) | |
| [3] | 张必东, 林泓, 朱思颖, 李忠成, 庄慧, 李云峰. 水稻颖壳异常突变体ah1的鉴定与候选基因分析[J]. 中国农业科学, 2024, 57(3): 429-441. |
| Zhang B D, Lin H, Zhu S Y, Li Z C, Zhuang H, Li Y F. Identification and candidate gene analysis of the ABNORMAL HULL 1 (ah1) mutant in rice (Oryza sativa L.)[J]. Scientia Agricultura Sinica, 2024, 57(3): 429-441. (in Chinese with English abstract) | |
| [4] | 罗曦, 魏林燕, 郑燕梅, 魏毅东, 连玲, 谢华安, 吴方喜. 水稻护颖发育相关基因的研究进展[J]. 南京农业大学学报2021, 44(3): 412-420. |
| Luo X, Wei L Y, Zheng Y M, Wei Y D, Lian L, Xie H A, Wu F X. Research advances on developmental genes of sterile lemmas in rice[J]. Journal of Nanjing Agricultural University, 2021, 44(3): 412-420. (in Chinese with English abstract) | |
| [5] | Ren D Y, Hu J. FZP determines grain size and sterile lemma fate in rice[J]. Journal of Experimental Botany, 2018, 69(20): 4853-4866. |
| [6] | Yoshida A, Suzaki T, Tanaka W, Hirano H Y. The homeotic gene long sterile lemma (G1) specifies sterile lemma identity in the rice spikelet[J]. Proceedings of the National Academy of Sciences of the United States of Ameica, 2009, 106 (47): 20103-20108. |
| [7] | Liu M, Li H, Su Y, Li W, Shi C. G1/ELE functions in the development of rice lemmas in addition to determining identities of empty glumes[J]. Frontiers in Plant Science, 2016, 7: 1006. |
| [8] | Kobayashi K, Maekawa M, Miyao A, Hirochika H, Kyozuka J. PANICLE PHYTOMER2 (PAP2), encoding a SEPALLATA subfamily MADS-box protein, positively controls spikelet meristem identity in rice[J]. Plant Cell Physiology, 2010, 51(1): 47-57. |
| [9] | Gao X, Liang W, Yin C, Ji S, Wang H, Su X, Guo C, Kong H, Xue H, Zhang D. The SEPALLATA-Like Gene OsMADS34 is required for rice inflorescence and spikelet development[J]. Plant Physiology, 2010, 153(2): 728-740. |
| [10] | Li W Q, Yoshida A, Takahashi M, Maekawa M, Kojima M, Sakakibara H, Kyozuka J. SAD1, an RNA polymerase Ⅰ subunit A34.5 of rice, interacts with mediator and controls various aspects of plant development[J]. The Plant Jouranl, 2015, 81: 282-291. |
| [11] | Xu Q K, Yu H P, Xia S S, Cui Y J, Yu X Q, Liu H, Zeng D L, Hu J, Zhang Q, Gao Z Y, Zhang G H, Zhu L, Shen L, Guo L B, Rao Y C, Qian Q, Ren D Y. The C2H2 zinc-finger protein LACKING RUDIMENTARY GLUME 1 regulates spikelet development in rice[J]. Science Bulletin, 2020, 65: 753-764. |
| [12] | Zhuang H, Wang H L, Zhang T, Zeng X Q, Chen H, Wang Z W, Zhang J, Zheng H, Tang J, Ling Y H, Yang Z L, He G H, Li Y F. NONSTOP GLUMES1 encodes a C2H2 zinc finger protein that regulates spikelet development in rice[J]. The Plant Cell, 2020, 32: 392-413. |
| [13] | Li H G, Xue D W, Gao Z Y, Yan M X, Xu W Y, Xing Z, Huang D N, Qian Q, Xue Y B. A putative lipase gene EXTRA GLUME1 regulates both empty-glume fate and spikelet development in rice[J]. The Plant Journal, 2009, 57(4): 593-605. |
| [14] | Yoshida A, Ohmori Y, Kitano H, Taguchi-Shiobara F, Hirano H Y. ABERRANT SPIKELET AND PANICLE1, encoding a TOPLESS-related transcriptional co-repressor, is involved in the regulation of meristem fate in rice[J]. The Plant Journal, 2012, 70(2): 327-339. |
| [15] | Yang J G, Lee S Y, Hang R L, Kim S R, Lee Y S, Cao X F, Amasino R, An G. OsVIL2 functions with PRC2 to induce flowering by repressing OsLFL1 in rice[J]. The Plant Journal, 2013, 73: 566-578. |
| [16] | Zhang J R, Tang W, Huang Y L, Niu X L, Zhao Y, Han Y, Liu Y S. Down-regulation of a LBD-like gene, OsIG1, leads to occurrence of unusual double ovules and developmental abnormalities of various floral organs and megagametophyte in rice[J]. Journal of Experimental Botany, 2015, 66(1): 99-112. |
| [17] | Lee D Y, Lee J, Moon S, Park S Y, An G. The rice heterochronic gene SUPERNUMERARY BRACT regulates the transition from spikelet meristem to floral meristem[J]. The Plant Journal, 2007, 49(1): 64-78. |
| [18] | Lee D Y, An G. Two AP2 family genes, SUPERNUMERARY BRACT (SNB) and OsINDETERMINATE SPIKELET1 (OsIDS1) synergistically control inflorescence architecture and floral meristem establishment in rice[J]. The Plant Journal, 2012, 69(3): 445-461. |
| [19] | Ren D Y, Li Y F, Zhao F M, Sang X C, Shi J Q, Wang N, Guo S, Ling Y H, Zhang C W, Yang Z L, He G H. MULTI-FLORET SPIKELET1, which encodes an AP2/ERF protein, determines spikelet meristem fate and sterile lemma identity in rice[J]. Plant Physiology, 2013, 162(2): 872-884. |
| [20] | Fan C C, Xing Y Z, Mao H L, Lu T T, Han B, Xu C G, Li X H, Zhang Q F. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein[J]. Theoretical and Applied Genetics, 2006, 112(6): 1164-1171. |
| [21] | Mao H L, Sun S Y, Yao J L, Wang C R, Yu S B, Xu C G, Li X H, Zhang Q F. Linking differential domain functions of the GS3 protein to natural variation of grain size in rice[J]. Proceedings of the National Academy of Sciences of the United States of USA, 2010, 107: 19579-19584. |
| [22] | Sun S Y, Wang L, Mao H L, Shao L, Li X H, Xiao J. H, Ouyang Y D, Zhang Q F. A G-protein pathway determines grain size in rice[J]. Nature Communications, 2018, 9(1): 851. |
| [23] | Qi P, Lin Y S, Song X J, Shen J B, Huang W, Shan J X, Zhu M Z, Jiang L, Gao J P, Lin H X. The novel quantitative trait locus GL3.1 controls rice grain size and yield by regulating Cyclin-T1;3[J]. Cell Research, 2012, 22: 1666-1680. |
| [24] | Zhang X J, Wang J F, Huang J, Lan H X, Wang C L, Yin C F, Wu Y Y, Tang H J, Qian Q, Li J Y, Zhang H S. Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice[J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109: 21534-21539. |
| [25] | Yu J P, Xiong H Y, Zhu X Y, Zhang H L, Li H H, Miao J L, Wang W S, Tang Z S, Zhang Z Y, Yao G X, Zhang Q, Pan Y H, Wang X, Rashid M A R, Li J J, Gao Y M, Li Z K, Yang W C, Fu X D, Li Z C. OsLG3 contributing to rice grain length and yield was mined by Ho-LAMap[J]. BMC Biology, 2017, 15: 28. |
| [26] | Ying J Z, Ma M, Bai C, Huang X H, Liu J L, Fan Y Y, Song X J. TGW3, a major QTL that negatively modulates grain length and weight in rice[J]. Molecular Plant, 2018, 11: 750-753. |
| [27] | Hu Z J, Lu S J, Wang M J, He H H, Sun L, Wang H R, Liu X H, Jiang L, Sun J L, Xin X Y, Kong W, Chu C C, Xue H W, Yang J S, Luo X J, Liu J X. A novel QTL qTGW3 encodes the GSK3/SHAGGY-Like kinase OsGSK5/OsSK41 that interacts with OsARF4 to negatively regulate grain size and weight in rice[J]. Molecular Plant, 2018, 11: 736-749. |
| [28] | Song X J, Kuroha T, Ayano M, Furuta T, Nagai K, Komeda N, Segami S, Miura K, Ogawa D, Kamura T, Suzuki T, Higashiyama T, Yamasaki M, Mori H, Inukai Y, Wu J Z, Kitano H, Sakakibara H, Jacobsen S E, Ashikari M. Rare allele of a previously unidentified histone H4 acetyltransferase enhances grain weight, yield, and plant biomass in rice[J]. Proceedings of the National Academy of Sciences USA, 2015, 112: 76-81. |
| [29] | Wang A H, Hou Q Q, Si L Z, Huang X H, Luo J H, Lu D F, Zhu J J, Shangguan Y Y, Miao J H, Xie Y F, Wang Y C, Zhao Q, Feng Q, Zhou C C, Li Y, Fan D L, Lu Y Q, Tian Q L, Wang Z X, Han B. The PLATZ transcription factor GL6 affects grain length and number in rice[J]. Plant Physiology, 2019, 180: 2077-2090. |
| [30] | Ishimaru K, Hirotsu N, Madoka Y, Murakami N, Hara N, Onodera H, Kashiwagi T, Ujiie K, Shimizu B I, Onishi A, Miyagawa H, Katoh E. Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield[J]. Nature Genetics, 2013, 45(6): 707-711. |
| [31] | Wang S K, Li S, Liu Q, Wu K, Zhang J Q, Wang S S, Wang Y, Chen X B, Zhang Y, Gao C X, Wang F, Huang H X, Fu X D. The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality[J]. Nature Genetics, 2015, 47: 949-954. |
| [32] | Wang Y X, Xiong G S, Hu J, Jiang L, Yu H, Xu J, Fang Y X, Zeng L J, Xu E B, Xu J, Ye W J, Meng X B, Liu R F, Chen H Q, Jing Y H, Wang Y H, Zhu X D, Li J Y, Qian Q. Copy number variation at the GL7 locus contributes to grain size diversity in rice[J]. Nature Genetics, 2015, 47: 944-948. |
| [33] | Si L Z, Chen J Y, Huang X H, Gong H, Luo J H, Hou Q Q, Zhou T Y, Lu T T, Zhu J J, Shangguan Y Y, Chen E W, Gong C X, Zhao Q, Jing Y F, Zhao Y, Li Y, Cui L L, Fan D L, Lu Y Q, Weng Q J, Wang Y C, Zhan Q L, Liu K Y, Wei X H, An K, An G, Han B. OsSPL13 controls grain size in cultivated rice[J]. Nature Genetics, 2016, 48: 447-456. |
| [34] | Song X J, Huang W, Shi M, Zhu M Z, Lin H X. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase[J]. Nature Genetics, 2007, 39: 623-630. |
| [35] | Hao J, Wang D, Wu Y, Huang K, Duan P G, Li N, Xu R, Zeng D L, Dong G J, Zhang B L, Zhang L M, Inzé D, Qian Q, Li Y H. The GW2-WG1-OsbZIP47 pathway controls grain size and weight in rice[J]. Molecular Plant, 2021, 8: 1266-1280. |
| [36] | Ruan B P, Shang L G, Zhang B, Hu J, Wang Y X, Lin H, Zhang A P, Liu C L, Peng Y L, Zhu L, Ren D Y, Shen L, Dong G J, Zhang G H, Zeng D L, Guo L B, Qian Q, Gao Z Y. Natural variation in the promoter of TGW2 determines grain width and weight in rice[J]. New Phytologist, 2020, 227: 629-640. |
| [37] | Li Y B, Fan C C, Xing Y Z, Jiang Y H, Luo L J, Sun L, Shao D, Xu C J, Li X H, Xiao J H, He Y Q, Zhang Q F. Natural variation in GS5 plays an important role in regulating grain size and yield in rice[J]. Nature Genetics, 2011, 43: 1266-1269. |
| [38] | Shomura A, Izawa T, Ebana K, Ebitani T, Kanegae H, Konishi S, Yano M. Deletion in a gene associated with grain size increased yields during rice domestication[J]. Nature Genetics, 2008, 40: 1023-1028. |
| [39] | Weng J F, Gu S H, Wan X Y, Gao H, Guo T, Su N, Lei C L, Zhang X, Cheng Z J, Guo X P, Wang J L, Jiang L, Zhai H Q, Wan J M. Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight[J]. Cell Research, 2008, 18: 1199-1209. |
| [40] | Duan P G, Xu J S, Zeng D L, Zhang B L, Geng M F, Zhang G Z, Huang K, Huang L J, Xu R, Ge S, Qian Q, Li Y H. Natural variation in the promoter of GSE5 contributes to grain size diversity in rice[J]. Molecular Plant, 2017, 10(5): 685-694. |
| [41] | Wang S K, Wu K, Yuan Q B, Liu X Y, Liu Z B, Lin X Y, Zeng R Z, Zhu H T, Dong G J, Qian Q, Zhang G Q, Fu X D. Control of grain size, shape and quality by OsSPL16 in rice[J]. Nature Genetics, 2012, 44: 950-954. |
| [42] | Rogers S O, Bendich A J. Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues[J]. Plant Molecular Biology, 1985, 5: 69-76. |
| [43] | Howarth W O. The Gramineae: A study of cereal, bamboo and grasses[J]. Nature, 1935, 136(3435): 317-319. |
| [44] | Kellogg E A. The evolutionary history of Ehrhartoideae, Oryzeae, and Oryza[J]. Rice, 2009, 2: 1-14. |
| [45] | Terrell E E, Peterson P M, Wergin W P. Epidermal features and spikelet micromorphology in Oryza and related genera (Poaceae: Oryzeae)[J]. Smithsonian Contributions to Botany, 2001, 91: 1-50. |
| [46] | Wu T K, Ali A, Wang J H, Song J H, Fang Y Q, Zhou T T, Luo Y, Zhang H Y, Chen X Q, Liao Y X, Liu Y T, Xu P Z, Wu J J. A homologous gene of OsREL2/ASP1, ASP-LSL regulates pleiotropic phenotype including long sterile lemma in rice[J]. BMC Plant Biology, 2021, 21: 390. |
| [1] | TAO Shibo, XU Na, XU Zhengjin, LIU Chang, XU Quan. Cloning of Cold6 Conferring Cold Tolerance in Rice [J]. Chinese Journal OF Rice Science, 2025, 39(6): 751-759. |
| [2] | HUANG Tao, WEI Zhaogen, CHENG Qi, CHENG Ze, LIU Xin, WANG Guangda, HU Keming, XIE Wenya, CHEN Zongxiang, FENG Zhiming, ZUO Shimin. Gene Cloning and Broad-spectrum Disease Resistance Analysis of Rice Lesion Mimic Mutant lm52 [J]. Chinese Journal OF Rice Science, 2025, 39(3): 322-330. |
| [3] | LIANG Chuyan, ZENG Wei, WANG Jiebing, YE Jing, WU Mingming, ZHAI Rongrong, ZHANG Xiaoming, ZHANG Hengmu, YE Shenghai. Characterization and Transcriptome Analysis of a Mutant with Short Panicle and Small Grain from Zhejing 99 [J]. Chinese Journal OF Rice Science, 2025, 39(1): 67-81. |
| [4] | HOU Xiaoqin, WANG Ying, YU Bei, FU Weimeng, FENG Baohua, SHEN Yichao, XIE Hangjun, WANG Huanran, XU Yongqiang, WU Zhihai, WANG Jianjun, TAO Longxing, FU Guanfu. Mechanisms Behind the Role of Potassium Fulvic Acid in Enhancing Salt Tolerance in Rice Seedlings [J]. Chinese Journal OF Rice Science, 2024, 38(4): 409-421. |
| [5] | LIU Huimin, ZHOU Jieqiang, HU Yuanyi, TIAN Yan, LEI Bin, LI Jianwu, WEI Zhongwei, TANG Wenbang. Super-high Yield Characteristics of Two-line Hybrid Rice Zhuoliangyou 1126 [J]. Chinese Journal OF Rice Science, 2024, 38(2): 160-171. |
| [6] | LIAN Yuanxun, WEI Ziyun, ZHANG Qiang, LI Qing, REN Deyong, HU Jiang, ZHU Li, GAO Zhenyu, ZHANG Guangheng, GUO Longbiao, ZENG Dali, QIAN Qian, SHEN Lan. Identification and Gene Mapping of a Zebra Leaf Mutant zl7 in Rice [J]. Chinese Journal OF Rice Science, 2023, 37(2): 113-124. |
| [7] | LIANG Cheng, XIANG Xunchao, ZHANG Ouling, YOU Hui, XU Liang, CHEN Yongjun. Analyses on Agronomic Traits and Genetic Characteristics of Two New Plant-architecture Lines in Rice [J]. Chinese Journal OF Rice Science, 2022, 36(2): 171-180. |
| [8] | Xiaojie CHU, Tao LU, Hanfei YE, Sheng WANG, Han LIN, Xianmei WU, Rui HE, Gang YAN, Yuexing WANG, Sanfeng LI, Mei LU, Haitao HU, Yaolong YANG, Yuchun RAO. Cloning and Functional Analysis of Leaf Senescence Gene LPS1 in Oryza sativa [J]. Chinese Journal OF Rice Science, 2021, 35(5): 427-438. |
| [9] | Yujun ZHU, Ziwei ZUO, Zhenhua ZHANG, Yeyang FAN. A New Approach for Fine-mapping and Map-based Cloning of Minor-Effect QTL in Rice [J]. Chinese Journal OF Rice Science, 2021, 35(4): 407-414. |
| [10] | Yali ZHENG, Linchuang YU, Xiaoxiao AN, Xinle CHENG, Lijun REN, Zilong SU, Xiaoya ZHENG, Tao LAN. Identification of a Knockout Mutant of OsWOX3B Gene in Rice (Oryza sativa L.) [J]. Chinese Journal OF Rice Science, 2021, 35(2): 112-120. |
| [11] | Yiwei KANG, Yuyu CHEN, Yingxin ZHANG. Research Progress and Breeding Prospects of Grain Size Associated Genes in Rice [J]. Chinese Journal OF Rice Science, 2020, 34(6): 479-490. |
| [12] | Tao XIONG, Yuanyuan NIE, Fangming MAO, Jianguo LEI, Linghua MAO, Shan ZHU, Renliang HUANG, Xianhua SHEN, Song YAN. Identification and Genetic Analysis of Cross-incompatibility ina RiceDW-type Sterile Line [J]. Chinese Journal OF Rice Science, 2020, 34(6): 520-524. |
| [13] | Wenbang TANG, Guilian ZHANG, Huabing DENG. Technology Exploration and Practice of Hybrid Rice Mechanized Seed Production [J]. Chinese Journal OF Rice Science, 2020, 34(2): 95-103. |
| [14] | Yanhua CHEN, Yaliang WANG, Defeng ZHU, Qinghua SHI, Huizhe CHEN, Jing XIANG, Yikai ZHANG, Yuping ZHANG. Mechanism of Exogenous Brassinolide in Alleviating High Temperature Injury at Panicle Initiation Stage in Rice [J]. Chinese Journal OF Rice Science, 2019, 33(5): 457-466. |
| [15] | Baoxuan NONG, Bixia QIN, Xiuzhong XIA, Xinghai YANG, Zongqiong ZHANG, Yu ZENG, Guofu DENG, Jianhe CAI, Zhanbiao LI, Piqing LIU, Danting LI. Genetic Analysis and Fine Mapping of a Major QTL for the Resistance to Southern Rice Black-Streaked Dwarf Disease [J]. Chinese Journal OF Rice Science, 2019, 33(2): 135-143. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||